New ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes in Wheat Backgrounds
Abstract
:1. Introduction
2. Results
2.1. Production of Thinopyrum Chromosome-Specific Oligo Probes
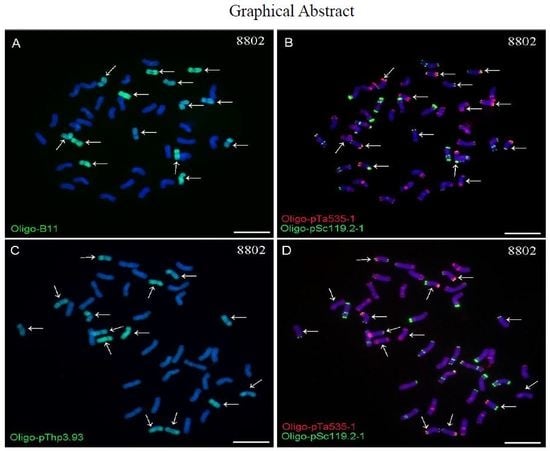
2.2. ND-FISH Analysis Using Oligo-B11 and Oligo-pThp3.93
3. Discussion
3.1. Using Oligo Probes and ND-FISH Assay to Identify Alien Chromosomes
3.2. ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes
4. Materials and Methods
4.1. Plant Materials
4.2. Cytological Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Han, F.; Liu, B.; Fedak, G.; Liu, Z. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 2004, 109, 1070–1076. [Google Scholar] [CrossRef]
- Fu, S.; Lv, Z.; Qi, B.; Guo, X.; Li, J.; Liu, B.; Han, F. Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium Head Blight. J. Genet. Genomics 2012, 39, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Tang, C.G.; Zhao, J.; Zheng, Q.; Li, B.; Hao, C.Y.; Li, Z.S.; Zhang, X.Y. Isolation of Thinopyrum ponticum genome specific repetitive sequences and their application for effective detection of alien segments in wheat. Sci. Agric. Sin. 2016, 49, 3683–3693. [Google Scholar]
- Li, J.; Lang, T.; Li, B.; Yu, Z.; Wang, H.; Li, G.; Yang, E.; Yang, Z. Introduction of Thinopyrum intermedium ssp. trichophorum chromosomes to wheat by trigeneric hybridization involving Triticum, Secale and Thinopyrum genera. Planta 2017, 245, 1121–1135. [Google Scholar] [CrossRef]
- Lang, T.; La, S.; Li, B.; Yu, Z.; Chen, Q.; Li, J.; Yang, E.; Li, G.; Yang, Z. Precise identification of wheat-Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 2018, 61, 177–185. [Google Scholar] [CrossRef]
- Liu, L.; Luo, Q.; Teng, W.; Li, B.; Li, H.; Li, Y.; Li, Z.; Zheng, Q. Development of Thinopyrum ponticum-specific molecular markers and FISH probes based on SLAF-seq technology. Planta 2018, 247, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, T.; Wu, Y.; Zhang, X.; Zhu, W.; Wang, Y.; Zeng, J.; Xu, L.; Fan, X.; Sha, L.; et al. FISH-based markers enable identification of chromosomes derived from tetraploid Thinopyrum elongatum in hybrid lines. Front. Plant Sci. 2018, 9, 526. [Google Scholar] [CrossRef] [PubMed]
- Mago, R.; Zhang, P.; Xia, X.; Zhang, J.; Hoxha, S.; Lagudah, E.; Graner, A.; Dundas, I. Transfer of stem rust resistance gene SrB from Thinopyrum ponticum into wheat and development of a closely linked PCR-based marker. Theor. Appl. Genet. 2019, 132, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Wang, J.; Lu, M.; Sun, S.; Chen, X.; Zhao, J.; Yang, Q.; Wu, J. Characterization of a wheat-Psathyrostachys huashanica Keng 4Ns disomic addition line for enhanced tiller numbers and stripe rust resistance. Planta 2014, 239, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Chen, L.; Wang, Y.; Li, M.; Yang, Z.; Qiu, L.; Yan, B.; Ren, Z.; Tang, Z. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef]
- Xiao, Z.; Tang, S.; Qiu, L.; Tang, Z.; Fu, S. Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 2017, 22, 973. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Golczyk, H.; Jouve, N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 2009, 1, 755–762. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Jouve, N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 2010, 119, 495–503. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Carmona, A.; Jouve, N. Chromosomal characterization of the three subgenomes in the polyploids of Hordeum murinum L.: New insight into the evolution of this complex. PLoS ONE 2013, 8, e81385. [Google Scholar] [CrossRef]
- Carmona, A.; Friero, E.; Bustos, A.D.; Jouve, N.; Cuadrado, A. Cytogenetic diversity of SSR motifs within and between Hordeum species carrying the H genome: H. vulgare L. and H. bulbosum L. Theor. Appl. Genet. 2013, 126, 949–961. [Google Scholar] [CrossRef]
- Cabo, S.; Carvalho, A.; Martin, A.; Lima-Brito, J. Structural rearrangements detected in newly-formed hexaploid tritordeum after three sequential FISH experiments with repetitive DNA sequences. J. Genet. 2014, 93, 183–188. [Google Scholar] [CrossRef] [Green Version]
- Delgado, A.; Carvalh, A.; Martín, A.C.; Martín, A.; Lima-Brito, J. Use of the synthetic Oligo-pTa535 and Oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid tritordeum. Genet. Resour. Crop Evol. 2016, 63, 945–951. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Lang, T.; Li, J.; La, S.; Yang, E.; Yang, Z. New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 2016, 244, 865–876. [Google Scholar] [CrossRef]
- Delgado, A.; Carvalho, A.; Martín, A.C.; Martín, A.; Lima-Brito, J. Genomic reshuffling in advanced lines of hexaploid tritordeum. Gene. Resour. Crop Evol. 2017, 64, 1331–1353. [Google Scholar] [CrossRef]
- Jiang, M.; Xiao, Z.Q.; Fu, S.L.; Tang, Z.X. FISH karyotype of 85 common wheat cultivars/lines displayed by ND-FISH using oligonucleotide probes. Cereal Res. Commun. 2017, 45, 549–563. [Google Scholar] [CrossRef]
- Duan, Q.; Wang, Y.Y.; Qiu, L.; Ren, T.H.; Li, Z.; Fu, S.L.; Tang, Z. Physical location of new PCR-based markers and powdery mildew resistance gene(s) on rye (Secale cereale L.) chromosome 4 using 4R dissection lines. Front. Plant Sci. 2017, 8, 1716. [Google Scholar] [CrossRef]
- Du, P.; Zhuang, L.; Wang, Y.; Yuan, L.; Wang, Q.; Wan, D.; Dawadondup; Tan, L.; Shen, J.; Xu, H.; Zhao, H.; et al. Development of oligonucleotides and multiplex probes for quick and accurate identitication of wheat and Thinopyrum bessarabicum chromosomes. Genome 2017, 60, 93–103. [Google Scholar] [CrossRef]
- Tang, S.; Tang, Z.; Qiu, L.; Yang, Z.; Li, G.; Lang, T.; Zhu, W.; Zhang, J.; Fu, S. Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef]
- Ren, T.; He, M.; Sun, Z.; Tan, F.; Luo, P.; Tang, Z.; Fu, S.; Yan, B.; Ren, Z.; Li, Z. The polymorphisms of oligonucleotide probes in wheat cultivars determined by ND-FISH. Molecules 2019, 24, 1126. [Google Scholar] [CrossRef]
- Lang, T.; Li, G.; Wang, H.; Yu, Z.; Chen, Q.; Yang, E.; Fu, S.; Tang, Z.; Yang, Z. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2019, 249, 663–675. [Google Scholar] [CrossRef]
- Du, H.; Tang, Z.; Duan, Q.; Tang, S.; Fu, S. Using the 6RLKu minichromosome of rye (Secale cereale L.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 2018, 19, E3933. [Google Scholar] [CrossRef]
- Nie, L.; Yang, Y.; Zhang, J.; Fu, T. Disomic chromosome addition from Thinopyrum intermedium to bread wheat appears to confer stripe rust resistance. Euphytica 2019, 215, 56. [Google Scholar] [CrossRef]
- Guo, X.; Shi, Q.; Wang, J.; Hou, Y.; Wang, Y.; Han, F. Characterization and genome changes of new amphiploids from wheat wide hybridization. J. Genet. Genomics 2015, 42, 459–461. [Google Scholar] [CrossRef]
- He, F.; Wang, Y.; Bao, Y.; Ma, Y.; Wang, X.; Li, X.; Wang, H. Chromosomal constitutions of five wheat–Elytrigia elongata partial amphiploids as revealed by GISH, multicolor GISH and FISH. Comp. Cytogenet. 2017, 11, 525–540. [Google Scholar] [CrossRef]
- Han, F.; Lamb, J.C.; Birchler, A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Yang, Z.; Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]




| Probe | Amount for Each Slide (ng/slide) | Oligonucleotide Sequence (5′-3′) |
|---|---|---|
| Oligo-B11 | 72.5–96.6 | TCCGCTCACCTTGATGACAACATCAGGTGGAATTCCGTTCGAGGG |
| Oligo-pThp3.93 | 69.5–92.6 | GGACTCCCACTAGATGTATCCGTCAAGGTGAATCCAGAGGAATCACCCTCGATGGCATT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, W.; Tang, Z.; Tang, S.; Yang, Z.; Luo, J.; Fu, S. New ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes in Wheat Backgrounds. Int. J. Mol. Sci. 2019, 20, 2031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20082031
Xi W, Tang Z, Tang S, Yang Z, Luo J, Fu S. New ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes in Wheat Backgrounds. International Journal of Molecular Sciences. 2019; 20(8):2031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20082031
Chicago/Turabian StyleXi, Wei, Zongxiang Tang, Shuyao Tang, Zujun Yang, Jie Luo, and Shulan Fu. 2019. "New ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes in Wheat Backgrounds" International Journal of Molecular Sciences 20, no. 8: 2031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20082031