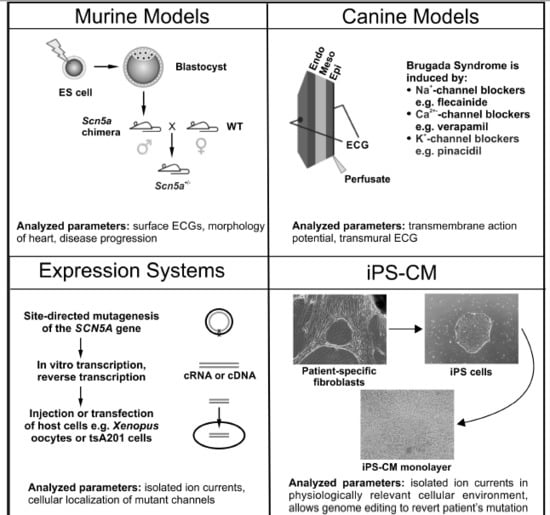
Experimental Models of Brugada syndrome
Abstract
:1. Introduction
2. Murine Models
2.1. Scn5a Heterozygous Knockout Mice
2.2. Scn5a-1798insD Knock-in Mice
2.3. Strengths and Limitations of Mouse Models
3. Porcine Model
4. Canine Models
4.1. Use of Canine Models to Uncover the Biophysical Causes of the Alterations in the Electrocardiogram
4.1.1. The J-wave/Osborne Wave
4.1.2. The ST-Segment Elevation
4.1.3. T-Wave Alternans and Ventricular Tachycardia or Fibrillation
4.2. Use of Canine Models to Assess the Cause for Sex Differences
4.3. Use of Canine Models to Identify Markers of Risk and to Improve Treatment
4.4. Strengths and Limitations of the Canine Model
5. Expression Systems
5.1. Use of Expression Systems to Assess the Biophysical Consequences of Brugada Syndrome-Associated Mutations
5.1.1. Sodium Current Density
5.1.2. Voltage Dependence of Activation and Steady State Inactivation
5.1.3. Inactivation Kinetics
5.1.4. Recovery from Inactivation
5.1.5. Other Genes Associated with Brugada Syndrome
5.2. Strengths and Limitations of Expression Systems
6. Induced Pluripotent Stem Cell-Derived Cardiomyocytes (iPS-CM)
Strengths and Limitations of iPS-CM
7. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| AV node | atrial ventricular node |
| HEK | human embryonic kidney |
| iPS cells | induced pluripotent stem cells |
| iPS-CM | induced pluripotent stem cell-derived cardiomyocytes |
| LQT syndrome | long QT syndrome |
| P2R | phase 2 reentry |
| INa | sodium inward current |
| tsA201 cells | immortalized HEK293 cells |
References
- Brugada, P.; Brugada, J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: A distinct clinical and electrocardiographic syndrome. A multicenter report. J. Am. Coll. Cardiol. 1992, 20, 1391–1396. [Google Scholar] [CrossRef]
- Chen, Q.; Kirsch, G.; Zhang, D.; Brugada, R.; Brugada, J.; Brugada, P.; Potenza, D.; Moya, A.; Borggrefe, M.; Breithardt, G.; et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature 1998, 392, 293–296. [Google Scholar] [CrossRef]
- Coronel, R.; Casini, S.; Koopmann, T.T.; Wilms-Schopman, F.J.; Verkerk, A.O.; de Groot, J.R.; Bhuiyan, Z.; Bezzina, C.R.; Veldkamp, M.W.; Linnenbank, A.C.; et al. Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome: A combined electrophysiological, genetic, histopathologic, and computational study. Circulation 2005, 112, 2769–2777. [Google Scholar] [CrossRef]
- Wilde, A.A.M.; Amin, A.S. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. JACC Clin. Electrophysiol. 2018, 4, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Brugada, J.; Brugada, R.; Brugada, P. Pharmacological and device approach to therapy of inherited cardiac diseases associated with cardiac arrhythmias and sudden death. J. Electrocardiol. 2000, 33, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Brugada, J.; Campuzano, O.; Arbelo, E.; Sarquella-Brugada, G.; Brugada, R. Present Status of Brugada Syndrome: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018, 72, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Minamino, T. Genetics of Brugada syndrome. J. Hum. Genet. 2016, 61, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Antzelevitch, C.; Pollevick, G.D.; Cordeiro, J.M.; Casis, O.; Sanguinetti, M.C.; Aizawa, Y.; Guerchicoff, A.; Pfeiffer, R.; Oliva, A.; Wollnik, B.; et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation 2007, 115, 442–449. [Google Scholar] [CrossRef]
- Cordeiro, J.M.; Marieb, M.; Pfeiffer, R.; Calloe, K.; Burashnikov, E.; Antzelevitch, C. Accelerated inactivation of the L-type calcium current due to a mutation in CACNB2b underlies Brugada syndrome. J. Mol. Cell. Cardiol. 2009, 46, 695–703. [Google Scholar] [CrossRef]
- London, B.; Michalec, M.; Mehdi, H.; Zhu, X.; Kerchner, L.; Sanyal, S.; Viswanathan, P.C.; Pfahnl, A.E.; Shang, L.L.; Madhusudanan, M.; et al. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation 2007, 116, 2260–2268. [Google Scholar] [CrossRef]
- Watanabe, H.; Koopmann, T.T.; Le Scouarnec, S.; Yang, T.; Ingram, C.R.; Schott, J.J.; Demolombe, S.; Probst, V.; Anselme, F.; Escande, D.; et al. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J. Clin. Investig. 2008, 118, 2260–2268. [Google Scholar]
- Riuró, H.; Beltran-Alvarez, P.; Tarradas, A.; Selga, E.; Campuzano, O.; Vergés, M.; Pagans, S.; Iglesias, A.; Brugada, J.; Brugada, P.; et al. A missense mutation in the sodium channel β2 subunit reveals SCN2B as a new candidate gene for Brugada syndrome. Hum. Mutat. 2013, 34, 961–966. [Google Scholar]
- Hu, D.; Barajas-Martinez, H.; Burashnikov, E.; Springer, M.; Wu, Y.; Varro, A.; Pfeiffer, R.; Koopmann, T.T.; Cordeiro, J.M.; Guerchicoff, A.; et al. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ. Cardiovasc. Genet. 2009, 2, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Delpón, E.; Cordeiro, J.M.; Núñez, L.; Thomsen, P.E.; Guerchicoff, A.; Pollevick, G.D.; Wu, Y.; Kanters, J.K.; Larsen, C.T.; Hofman-Bang, J.; et al. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circ. Arrhythm. Electrophysiol. 2008, 1, 209–218. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Kim, R.; Udupa, S.; Costain, G.; Jobling, R.; Liston, E.; Jamal, S.M.; Szybowska, M.; Morel, C.F.; Bowdin, S.; et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death. Circulation 2018, 138, 1195–1205. [Google Scholar] [CrossRef]
- Yan, G.X.; Antzelevitch, C. Cellular basis for the Brugada syndrome and other mechanisms of arrhythmogenesis associated with ST-segment elevation. Circulation 1999, 100, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Meregalli, P.G.; Wilde, A.A.; Tan, H.L. Pathophysiological mechanisms of Brugada syndrome: Depolarization disorder, repolarization disorder, or more? Cardiovasc. Res. 2005, 67, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.A.; Postema, P.G.; Di Diego, J.M.; Viskin, S.; Morita, H.; Fish, J.M.; Antzelevitch, C. The pathophysiological mechanism underlying Brugada syndrome: Depolarization versus repolarization. J. Mol. Cell. Cardiol. 2010, 49, 543–553. [Google Scholar] [CrossRef]
- Nademanee, K.; Veerakul, G.; Chandanamattha, P.; Chaothawee, L.; Ariyachaipanich, A.; Jirasirirojanakorn, K.; Likittanasombat, K.; Bhuripanyo, K.; Ngarmukos, T. Prevention of Ventricular Fibrillation Episodes in Brugada Syndrome by Catheter Ablation over the Anterior Right Ventricular Outflow Tract Epicardium. Circulation 2011, 123, 1270–1279. [Google Scholar] [CrossRef]
- Szel, T.; Antzelevitch, C. Abnormal repolarization as the basis for late potentials and fractionated electrograms recorded from epicardium in experimental models of Brugada syndrome. J. Am. Coll. Cardiol. 2014, 63, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Tukkie, R.; Sogaard, P.; Vleugels, J.; de Groot, I.K.; Wilde, A.A.; Tan, H.L. Delay in right ventricular activation contributes to Brugada syndrome. Circulation 2004, 109, 1272–1277. [Google Scholar] [CrossRef]
- Papadatos, G.; Wallerstein, P.; Head, C.; Ratcliff, R.; Brady, P.; Benndorf, K.; Saumarez, R.; Trezise, A.; Huang, C.; Vandenberg, J.; et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl. Acad. Sci. USA 2002, 99, 6210–6215. [Google Scholar] [CrossRef] [PubMed]
- Leoni, A.; Gavillet, B.; Rougier, J.; Marionneau, C.; Probst, V.; Le Scouarnec, S.; Schott, J.; Demolombe, S.; Bruneval, P.; Huang, C.; et al. Variable Na(v)1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a(+/-) mouse model. PLoS ONE 2010, 5, 9298. [Google Scholar] [CrossRef]
- Jeevaratnam, K.; Poh Tee, S.; Zhang, Y.; Rewbury, R.; Guzadhur, L.; Duehmke, R.; Grace, A.A.; Lei, M.; Huang, C.L. Delayed conduction and its implications in murine Scn5a(+/-) hearts: Independent and interacting effects of genotype, age, and sex. Pflug. Arch. Eur. J. Physiol. 2011, 461, 29–44. [Google Scholar] [CrossRef]
- Jeevaratnam, K.; Rewbury, R.; Zhang, Y.; Guzadhur, L.; Grace, A.A.; Lei, M.; Huang, C.L. Frequency distribution analysis of activation times and regional fibrosis in murine Scn5a+/- hearts: The effects of ageing and sex. Mech. Ageing Dev. 2012, 133, 591–599. [Google Scholar] [CrossRef]
- Jeevaratnam, K.; Zhang, Y.; Guzadhur, L.; Duehmke, R.M.; Lei, M.; Grace, A.A.; Huang, C.L. Differences in sino-atrial and atrio-ventricular function with age and sex attributable to the Scn5a+/- mutation in a murine cardiac model. Acta Physiol. (Oxf) 2010, 200, 23–33. [Google Scholar]
- Royer, A.; van Veen, T.A.; Le Bouter, S.; Marionneau, C.; Griol-Charhbili, V.; Léoni, A.L.; Steenman, M.; van Rijen, H.V.; Demolombe, S.; Goddard, C.A.; et al. Mouse model of SCN5A-linked hereditary Lenègre’s disease: Age-related conduction slowing and myocardial fibrosis. Circulation 2005, 111, 1738–1746. [Google Scholar] [CrossRef]
- van Veen, T.A.; van Rijen, H.V.; van Kempen, M.J.; Miquerol, L.; Opthof, T.; Gros, D.; Vos, M.A.; Jongsma, H.J.; de Bakker, J.M. Discontinuous conduction in mouse bundle branches is caused by bundle-branch architecture. Circulation 2005, 112, 2235–2244. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y.; Grace, A.; Huang, C. Increased Right Ventricular Repolarization Gradients Promote Arrhythmogenesis in a Murine Model of Brugada Syndrome. J. Cardiovasc. Electrophysiol. 2010, 21, 1153–1159. [Google Scholar] [CrossRef]
- Matthews, G.D.; Martin, C.A.; Grace, A.A.; Zhang, Y.; Huang, C.L. Regional variations in action potential alternans in isolated murine Scn5a (+/-) hearts during dynamic pacing. Acta Physiol. (Oxf) 2010, 200, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Stokoe, K.; Balasubramaniam, R.; Goddard, C.; Colledge, W.; Grace, A.; Huang, C. Effects of flecainide and quinidine on arrhythmogenic properties of Scn5a+/- murine hearts modelling the Brugada syndrome. J. Physiol. 2007, 581, 255–275. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y.; Grace, A.; Huang, C. In vivo studies of Scn5a+/- mice modeling Brugada syndrome demonstrate both conduction and repolarization abnormalities. J. Electrocardiol. 2010, 43, 433–439. [Google Scholar] [CrossRef]
- Martin, C.A.; Siedlecka, U.; Kemmerich, K.; Lawrence, J.; Cartledge, J.; Guzadhur, L.; Brice, N.; Grace, A.A.; Schwiening, C.; Terracciano, C.M.; et al. Reduced Na(+) and higher K(+) channel expression and function contribute to right ventricular origin of arrhythmias in Scn5a+/- mice. Open Biol. 2012, 2, 120072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guzadhur, L.; Jeevaratnam, K.; Salvage, S.C.; Matthews, G.D.; Lammers, W.J.; Lei, M.; Huang, C.L.; Fraser, J.A. Arrhythmic substrate, slowed propagation and increased dispersion in conduction direction in the right ventricular outflow tract of murine Scn5a+/- hearts. Acta Physiol. (Oxf) 2014, 211, 559–573. [Google Scholar] [CrossRef]
- Huang, C.L. Murine Electrophysiological Models of Cardiac Arrhythmogenesis. Physiol. Rev. 2017, 97, 283–409. [Google Scholar] [CrossRef]
- Tse, G.; Wong, S.T.; Tse, V.; Yeo, J.M. Depolarization vs. repolarization: What is the mechanism of ventricular arrhythmogenesis underlying sodium channel haploinsufficiency in mouse hearts? Acta Physiol. (Oxf) 2016, 218, 234–235. [Google Scholar] [CrossRef] [PubMed]
- Tse, G.; Wong, S.T.; Tse, V.; Yeo, J.M. Determination of action potential wavelength restitution in Scn5a(+/-) mouse hearts modelling human Brugada syndrome. J. Geriatr. Cardiol. 2017, 14, 595–596. [Google Scholar]
- Kelly, A.; Salerno, S.; Connolly, A.; Bishop, M.; Charpentier, F.; Stolen, T.; Smith, G.L. Normal interventricular differences in tissue architecture underlie right ventricular susceptibility to conduction abnormalities in a mouse model of Brugada syndrome. Cardiovasc. Res. 2018, 114, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Remme, C.A.; Verkerk, A.O.; Nuyens, D.; van Ginneken, A.C.; van Brunschot, S.; Belterman, C.N.; Wilders, R.; van Roon, M.A.; Tan, H.L.; Wilde, A.A.; et al. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation 2006, 114, 2584–2594. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.P.; Casini, S.; van den Berg, C.W.; Hoekstra, M.; Remme, C.A.; Dambrot, C.; Salvatori, D.; Oostwaard, D.W.; Wilde, A.A.; Bezzina, C.R.; et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 2012, 125, 3079–3091. [Google Scholar] [CrossRef]
- Stein, M.; van Veen, T.A.; Remme, C.A.; Boulaksil, M.; Noorman, M.; van Stuijvenberg, L.; van der Nagel, R.; Bezzina, C.R.; Hauer, R.N.; de Bakker, J.M.; et al. Combined reduction of intercellular coupling and membrane excitability differentially affects transverse and longitudinal cardiac conduction. Cardiovasc. Res. 2009, 83, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, T.; Haufe, V.; Blechschmidt, S. Voltage-gated sodium channels in the mammalian heart. Glob. Cardiol. Sci. Pract. 2014, 2014, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Antzelevitch, C. Cellular basis for the electrocardiographic J wave. Circulation 1996, 93, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef]
- Kimura, M.; Kobayashi, T.; Owada, S.; Ashikaga, K.; Higuma, T.; Sasaki, S.; Iwasa, A.; Motomura, S.; Okumura, K. Mechanism of ST elevation and ventricular arrhythmias in an experimental Brugada syndrome model. Circulation 2004, 109, 125–131. [Google Scholar] [CrossRef]
- Szél, T.; Koncz, I.; Antzelevitch, C. Cellular mechanisms underlying the effects of milrinone and cilostazol to suppress arrhythmogenesis associated with Brugada syndrome. Heart Rhythm 2013, 10, 1720–1727. [Google Scholar] [CrossRef]
- Take, Y.; Morita, H.; Wu, J.; Nagase, S.; Morita, S.; Toh, N.; Nishii, N.; Nakamura, K.; Kusano, K.F.; Ohe, T.; et al. Spontaneous electrocardiogram alterations predict ventricular fibrillation in Brugada syndrome. Heart Rhythm 2011, 8, 1014–1021. [Google Scholar] [CrossRef]
- Nishida, K.; Fujiki, A.; Mizumaki, K.; Sakabe, M.; Sugao, M.; Tsuneda, T.; Inoue, H. Canine model of Brugada syndrome using regional epicardial cooling of the right ventricular outflow tract. J. Cardiovasc. Electrophysiol. 2004, 15, 936–941. [Google Scholar] [CrossRef]
- Morita, H.; Zipes, D.; Fukushima-Kusano, K.; Nagase, S.; Nakamura, K.; Morita, S.; Ohe, T.; Wu, J. Repolarization heterogeneity in the right ventricular outflow tract: Correlation with ventricular arrhythmias in Brugada patients and in an in vitro canine Brugada model. Heart Rhythm 2008, 5, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Zipes, D.; Lopshire, J.; Morita, S.; Wu, J. T wave alternans in an in vitro canine tissue model of Brugada syndrome. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, 421–428. [Google Scholar] [CrossRef]
- Morita, H.; Zipes, D.; Morita, S.; Wu, J. Genotype-phenotype correlation in tissue models of Brugada syndrome simulating patients with sodium and calcium channelopathies. Heart Rhythm 2010, 7, 820–827. [Google Scholar] [CrossRef]
- Hong, K.; Brugada, J.; Oliva, A.; Berruezo-Sanchez, A.; Potenza, D.; Pollevick, G.D.; Guerchicoff, A.; Matsuo, K.; Burashnikov, E.; Dumaine, R.; et al. Value of electrocardiographic parameters and ajmaline test in the diagnosis of Brugada syndrome caused by SCN5A mutations. Circulation 2004, 110, 3023–3027. [Google Scholar] [CrossRef]
- Fish, J.; Antzelevitch, C. Cellular and ionic basis for the sex-related difference in the manifestation of the Brugada syndrome and progressive conduction disease phenotypes. J. Electrocardiol. 2003, 36, 173–179. [Google Scholar] [CrossRef]
- Morita, H.; Kusano, K.; Miura, D.; Nagase, S.; Nakamura, K.; Morita, S.; Ohe, T.; Zipes, D.; Wu, J. Fragmented QRS as a marker of conduction abnormality and a predictor of prognosis of Brugada syndrome. Circulation 2008, 118, 1697–1704. [Google Scholar] [CrossRef]
- Morita, H.; Zipes, D.; Morita, S.; Lopshire, J.; Wu, J. Epicardial ablation eliminates ventricular arrhythmias in an experimental model of Brugada syndrome. Heart Rhythm 2009, 6, 665–671. [Google Scholar] [CrossRef]
- Patocskai, B.; Yoon, N.; Antzelevitch, C. Mechanisms Underlying Epicardial Radiofrequency Ablation to Suppress Arrhythmogenesis in Experimental Models of Brugada Syndrome. JACC. Clin. Electrophysiol. 2017, 3, 353–363. [Google Scholar] [CrossRef]
- Denham, N.C.; Pearman, C.M.; Ding, W.Y.; Waktare, J.; Gupta, D.; Snowdon, R.; Hall, M.; Cooper, R.; Modi, S.; Todd, D.; et al. Systematic re-evaluation of SCN5A variants associated with Brugada syndrome. J. Cardiovasc. Electrophysiol. 2019, 30, 118–127. [Google Scholar] [CrossRef]
- Baroudi, G.; Pouliot, V.; Denjoy, I.; Guicheney, P.; Shrier, A.; Chahine, M. Novel mechanism for Brugada syndrome: Defective surface localization of an SCN5A mutant (R1432G). Circ. Res. 2001, 88, 78–83. [Google Scholar] [CrossRef]
- Baroudi, G.; Napolitano, C.; Priori, S.G.; Del Bufalo, A.; Chahine, M. Loss of function associated with novel mutations of the SCN5A gene in patients with Brugada syndrome. Can. J. Cardiol. 2004, 20, 425–430. [Google Scholar]
- Shin, D.J.; Kim, E.; Park, S.B.; Jang, W.C.; Bae, Y.; Han, J.; Jang, Y.; Joung, B.; Lee, M.H.; Kim, S.S.; et al. A novel mutation in the SCN5A gene is associated with Brugada syndrome. Life Sci. 2007, 80, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Baroudi, G.; Chahine, M. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 2000, 487, 224–228. [Google Scholar] [CrossRef]
- Bezzina, C.; Veldkamp, M.W.; van Den Berg, M.P.; Postma, A.V.; Rook, M.B.; Viersma, J.W.; van Langen, I.M.; Tan-Sindhunata, G.; Bink-Boelkens, M.T.; van Der Hout, A.H.; et al. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 1999, 85, 1206–1213. [Google Scholar] [CrossRef]
- Tarradas, A.; Selga, E.; Beltran-Alvarez, P.; Perez-Serra, A.; Riuro, H.; Pico, F.; Iglesias, A.; Campuzano, O.; Castro-Urda, V.; Fernandez-Lozano, I.; et al. A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes Brugada syndrome. PLoS ONE 2013, 8, 53220. [Google Scholar] [CrossRef]
- Keller, D.I.; Rougier, J.S.; Kucera, J.P.; Benammar, N.; Fressart, V.; Guicheney, P.; Madle, A.; Fromer, M.; Schlapfer, J.; Abriel, H. Brugada syndrome and fever: Genetic and molecular characterization of patients carrying SCN5A mutations. Cardiovasc. Res. 2005, 67, 510–519. [Google Scholar] [CrossRef]
- Wan, X.; Chen, S.; Sadeghpour, A.; Wang, Q.; Kirsch, G.E. Accelerated inactivation in a mutant Na(+) channel associated with idiopathic ventricular fibrillation. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, 354–360. [Google Scholar] [CrossRef]
- Keller, D.I.; Barrane, F.Z.; Gouas, L.; Martin, J.; Pilote, S.; Suarez, V.; Osswald, S.; Brink, M.; Guicheney, P.; Schwick, N.; et al. A novel nonsense mutation in the SCN5A gene leads to Brugada syndrome and a silent gene mutation carrier state. Can. J. Cardiol. 2005, 21, 925–931. [Google Scholar]
- Vatta, M.; Dumaine, R.; Antzelevitch, C.; Brugada, R.; Li, H.; Bowles, N.E.; Nademanee, K.; Brugada, J.; Brugada, P.; Towbin, J.A. Novel mutations in domain I of SCN5A cause Brugada syndrome. Mol. Genet. Metab. 2002, 75, 317–324. [Google Scholar] [CrossRef]
- Rook, M.; Bezzina Alshinawi, C.; Groenewegen, W.; van Gelder, I.; van Ginneken, A.; Jongsma, H.; Mannens, M.; Wilde, A. Human SCN5A gene mutations alter cardiac sodium channel kinetics and are associated with the Brugada syndrome. Cardiovasc. Res. 1999, 44, 507–517. [Google Scholar] [CrossRef]
- Veldkamp, M.W.; Viswanathan, P.C.; Bezzina, C.; Baartscheer, A.; Wilde, A.A.; Balser, J.R. Two distinct congenital arrhythmias evoked by a multidysfunctional Na(+) channel. Circ. Res. 2000, 86, 91–97. [Google Scholar] [CrossRef]
- Makita, N.; Shirai, N.; Wang, D.W.; Sasaki, K.; George, A.L., Jr.; Kanno, M.; Kitabatake, A. Cardiac Na(+) channel dysfunction in Brugada syndrome is aggravated by beta(1)-subunit. Circulation 2000, 101, 54–60. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flügel, L.; Dorn, T.; Goedel, A.; Höhnke, C.; Hofmann, F.; et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef]
- Matsa, E.; Rajamohan, D.; Dick, E.; Young, L.; Mellor, I.; Staniforth, A.; Denning, C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur. Heart J. 2011, 32, 952–962. [Google Scholar] [CrossRef]
- Zhang, X.H.; Haviland, S.; Wei, H.; Saric, T.; Fatima, A.; Hescheler, J.; Cleemann, L.; Morad, M. Ca2+ signaling in human induced pluripotent stem cell-derived cardiomyocytes (iPS-CM) from normal and catecholaminergic polymorphic ventricular tachycardia (CPVT)-afflicted subjects. Cell Calcium 2013, 54, 57–70. [Google Scholar] [CrossRef]
- Yazawa, M.; Hsueh, B.; Jia, X.; Pasca, A.M.; Bernstein, J.A.; Hallmayer, J.; Dolmetsch, R.E. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011, 471, 230–234. [Google Scholar] [CrossRef]
- Kosmidis, G.; Veerman, C.C.; Casini, S.; Verkerk, A.O.; van de Pas, S.; Bellin, M.; Wilde, A.A.; Mummery, C.L.; Bezzina, C.R. Readthrough-Promoting Drugs Gentamicin and PTC124 Fail to Rescue Nav1.5 Function of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Carrying Nonsense Mutations in the Sodium Channel Gene SCN5A. Circ. Arrhythm. Electrophysiol. 2016, 9, e004227. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Sallam, K.; Wu, H.; Li, Y.; Itzhaki, I.; Garg, P.; Zhang, Y.; Vermglinchan, V.; Lan, F.; Gu, M.; et al. Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome. J. Am. Coll. Cardiol. 2016, 68, 2086–2096. [Google Scholar] [CrossRef]
- Veerman, C.C.; Mengarelli, I.; Guan, K.; Stauske, M.; Barc, J.; Tan, H.L.; Wilde, A.A.; Verkerk, A.O.; Bezzina, C.R. hiPSC-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci. Rep. 2016, 6, 30967. [Google Scholar] [CrossRef]
- Miller, D.C.; Harmer, S.C.; Poliandri, A.; Nobles, M.; Edwards, E.C.; Ware, J.S.; Sharp, T.V.; McKay, T.R.; Dunkel, L.; Lambiase, P.D.; et al. Ajmaline blocks INa and IKr without eliciting differences between Brugada syndrome patient and control human pluripotent stem cell-derived cardiac clusters. Stem Cell Res. 2017, 25, 233–244. [Google Scholar] [CrossRef]
- Doss, M.X.; Di Diego, J.M.; Goodrow, R.J.; Wu, Y.; Cordeiro, J.M.; Nesterenko, V.V.; Barajas-Martinez, H.; Hu, D.; Urrutia, J.; Desai, M.; et al. Maximum diastolic potential of human induced pluripotent stem cell-derived cardiomyocytes depends critically on I(Kr). PLoS ONE 2012, 7, 40288. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Liu, Z.; Loh, L.J.; Zhao, Y.; Li, G.; Liew, R.; Islam, O.; Wu, J.; Chung, Y.Y.; Teo, W.S.; et al. Identification of an INa-dependent and Ito-mediated proarrhythmic mechanism in cardiomyocytes derived from pluripotent stem cells of a Brugada syndrome patient. Sci. Rep. 2018, 8, 11246. [Google Scholar] [CrossRef] [PubMed]
- Selga, E.; Sendfeld, F.; Martinez-Moreno, R.; Medine, C.N.; Tura-Ceide, O.; Wilmut, S.I.; Perez, G.J.; Scornik, F.S.; Brugada, R.; Mills, N.L. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J. Mol. Cell. Cardiol. 2018, 114, 10–19. [Google Scholar] [CrossRef]
- Veerman, C.C.; Kosmidis, G.; Mummery, C.L.; Casini, S.; Verkerk, A.O.; Bellin, M. Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev. 2015, 24, 1035–1052. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Ma, J.; Guo, L.; Fiene, S.J.; Anson, B.D.; Thomson, J.A.; Kamp, T.J.; Kolaja, K.L.; Swanson, B.J.; January, C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, 2006–2017. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, J.; Han, P.; Yuan, Q.; Zhang, J.; Zhang, X.; Xu, Y.; Cao, H.; Meng, Q.; Chen, L.; et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011, 21, 579–587. [Google Scholar] [CrossRef]
- Uosaki, H.; Fukushima, H.; Takeuchi, A.; Matsuoka, S.; Nakatsuji, N.; Yamanaka, S.; Yamashita, J.K. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS ONE 2011, 6, e23657. [Google Scholar] [CrossRef] [PubMed]
- Le Scouarnec, S.; Karakachoff, M.; Gourraud, J.B.; Lindenbaum, P.; Bonnaud, S.; Portero, V.; Duboscq-Bidot, L.; Daumy, X.; Simonet, F.; Teusan, R.; et al. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum. Mol. Genet. 2015, 24, 2757–2763. [Google Scholar] [CrossRef]
- Bezzina, C.R.; Barc, J.; Mizusawa, Y.; Remme, C.A.; Gourraud, J.-B.; Simonet, F.; Verkerk, A.O.; Schwartz, P.J.; Crotti, L.; Dagradi, F.; et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 2013, 45, 1044. [Google Scholar] [CrossRef]
- Park, D.S.; Cerrone, M.; Morley, G.; Vasquez, C.; Fowler, S.; Liu, N.; Bernstein, S.A.; Liu, F.Y.; Zhang, J.; Rogers, C.S.; et al. Genetically engineered SCN5A mutant pig hearts exhibit conduction defects and arrhythmias. J. Clin. Invest. 2015, 125, 403–412. [Google Scholar] [CrossRef] [PubMed]



| Model | Major Findings | Advantages | Disadvantages |
|---|---|---|---|
| Murine | SCN5A+/−mouse:
|
|
|
| Canine |
|
|
|
| Porcine |
|
| |
| Heterologous expression | See Supplementary Table |
|
|
| iPS-CM |
|
|
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sendfeld, F.; Selga, E.; Scornik, F.S.; Pérez, G.J.; Mills, N.L.; Brugada, R. Experimental Models of Brugada syndrome. Int. J. Mol. Sci. 2019, 20, 2123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20092123
Sendfeld F, Selga E, Scornik FS, Pérez GJ, Mills NL, Brugada R. Experimental Models of Brugada syndrome. International Journal of Molecular Sciences. 2019; 20(9):2123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20092123
Chicago/Turabian StyleSendfeld, Franziska, Elisabet Selga, Fabiana S. Scornik, Guillermo J. Pérez, Nicholas L. Mills, and Ramon Brugada. 2019. "Experimental Models of Brugada syndrome" International Journal of Molecular Sciences 20, no. 9: 2123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20092123