Light- and Temperature-Induced Expression of an R2R3-MYB Gene Regulates Anthocyanin Biosynthesis in Red-Fleshed Kiwifruit
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification, Genomic Location, Gene Structure, and Motif Composition of R2R3-MYB Genes in Kiwifruit
2.2. Phylogenetic Analysis of AccR2R3-MYB TFs
2.3. Expression Preference of AccR2R3-MYBs in Kiwifruit
2.4. Identification of Cis-Acting Regulatory Elements of Anthocyanin-Related AccR2R3-MYBs
2.5. Light and Low-Temperature Synergistically Induced the Red Pigmentation in the Callus of A. chinensis cv. Hongyang
2.6. Sequence Analysis, Transcriptional Activation Ability, and Expression Characteristic of Acc00493 (AcMYB10)
2.7. Gene Functional Study by Overexpression of AcMYB10 in A. chinensis cv. Hongyang and Using the VIGS (Virus-Induced Gene Silencing) System in A. chinensis cv. Hongshi No. 2
2.8. Subcellular Localization Analysis
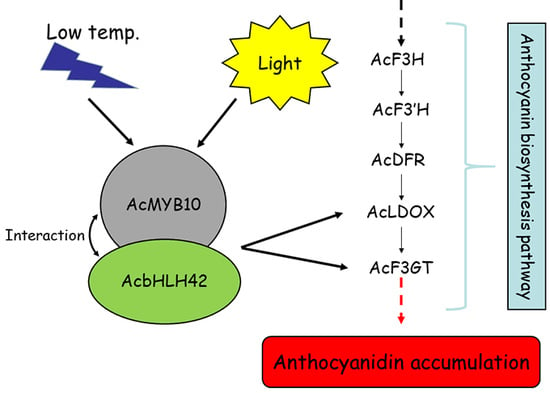
2.9. Protein–Protein Interactions between AcMYB10 and AcbHLH42
2.10. AcMYB10 and AcbHLH42 Regulate the Promoter Activities of AcLDOX and AcF3GT
2.11. AcMYB10 Interacts with AcbHLH42 to Activate Anthocyanin Biosynthesis in N. tabacum Leafs and A. arguta Fruit
3. Materials and Methods
3.1. Plant Materials, Treatment, and Nucleic Acid Extraction
3.2. Genome-Wide Analysis and Expression Profile of Kiwifruit R2R3-MYB Genes
3.3. Quantitative Real-Time RT-PCR
3.4. Determining the Concentration of Anthocyanin, Chlorophyll, and Flavonoid Metabolites
3.5. Isolation and Sequence Alignments and Transcriptional Activation Analysis of AcMYB10
3.6. Dual Luciferase Assay of Transiently Transformed Tobacco Leaves
3.7. Gene Functional Study Using the VIGS System in Kiwifruit and Over-Expression of AcMYB10 in A. chinensis, A. arguta, and N. tabacum
3.8. Subcellular Localization Analysis
3.9. Yeast Two-Hybrid (Y2H) Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HMM | Hidden Markov Model |
| CDS | coding sequences |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| VIGS | Virus-induced gene silencing |
| qRT-PCR | Quantitative real-time PCR |
| DAP | days after pollination |
| YFP | yellow fluorescent protein |
| RFP | red fluorescent protein |
| 3AT | 3-amino-1,2,4-triazole |
| CRE | Cis-acting regulatory element |
| LTR | Low-temperature stress |
| MRE | MYB-recognizing element |
References
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.L.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Du, H.; Yang, S.-S.; Liang, Z.; Feng, B.-R.; Liu, L.; Huang, Y.-B.; Tang, Y.-X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Pazares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea-mays encodes a protein with homology to myb protooncogene products and with structural similarities to transcriptional activators. Embo J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.-M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, X.; Soejarto, D. Actinidiaceae. Flora China 2007, 12, 334–360. [Google Scholar]
- Montefiori, M.; Comeskey, D.J.; Wohlers, M.; McGhie, T.K. Characterization and Quantification of Anthocyanins in Red Kiwifruit (Actinidia spp.). J. Agric. Food Chem. 2009, 57, 6856–6861. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.R.; Harker, F.R. Consumer evaluation of novel kiwifruit: Willingness-to-pay. J. Sci. Food Agric. 2005, 85, 2519–2526. [Google Scholar] [CrossRef]
- Yousuf, B.; Gul, K.; Wani, A.A.; Singh, P. Health Benefits of Anthocyanins and Their Encapsulation for Potential Use in Food Systems: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2223–2230. [Google Scholar] [CrossRef]
- Montefiori, M.; Espley, R.V.; Stevenson, D.; Cooney, J.; Datson, P.M.; Saiz, A.; Atkinson, R.G.; Hellens, R.P.; Allan, A.C. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2011, 65, 106–118. [Google Scholar] [CrossRef]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genom. 2013, 14, 28. [Google Scholar] [CrossRef]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 16861. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, B.; Qi, Y.; Chen, X.; Liu, C.; Liu, Z.; Ren, X. Expression Differences of Pigment Structural Genes and Transcription Factors Explain Flesh Coloration in Three Contrasting Kiwifruit Cultivars. Front. Plant Sci. 2017, 8, 1507. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. Cell Mol. Biol. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Li, X.-J.; Hu, B.; Qin, Y.-H.; Huang, X.-M.; Wang, H.-C.; Hu, G.-B. LcMYB1 Is a Key Determinant of Differential Anthocyanin Accumulation among Genotypes, Tissues, Developmental Phases and ABA and Light Stimuli in Litchi chinensis. PloS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Niu, S.-S.; Xu, C.-J.; Zhang, W.-S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.-S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Azuma, A.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012, 236, 1067–1080. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, Z.; Jiang, S.; Xu, H.; Wang, Y.; Feng, S.; Chen, X. Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ. Cult. 2016, 127, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Xia, Y.; Wang, Y.; Qin, G.; Tian, S. Characterization of Genes Encoding Key Enzymes Involved in Anthocyanin Metabolism of Kiwifruit during Storage Period. Front. Plant Sci. 2017, 8, 341. [Google Scholar] [CrossRef] [Green Version]
- Man, Y.-P.; Wang, Y.-C.; Li, Z.-Z.; Jiang, Z.-W.; Yang, H.-L.; Gong, J.-J.; He, S.-S.; Wu, S.-Q.; Yang, Z.-Q.; Zheng, J.; et al. High-temperature inhibition of biosynthesis and transportation of anthocyanins results in the poor red coloration in red-fleshed Actinidia chinensis. Physiol. Plant 2015, 153, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Yamane, T.; Jeong, S.T.; Goto-Yamamoto, N.; Koshita, Y.; Kobayashi, S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am. J. Enol. Vitic. 2006, 57, 54–59. [Google Scholar]
- Mohanty, S.; Grimm, B.; Tripathy, B.C. Light and dark modulation of chlorophyll biosynthetic genes in response to temperature. Planta 2006, 224, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.A.; McGhie, T.K.; White, A.; Woolf, A.B. Skin colour and pigment changes during ripening of ‘Haas’ avocado fruit. Postharvest Biol. Technol. 2004, 31, 287–294. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Li, J.G.; Yang, X.Y.; Wang, Y.; Li, X.J.; Gao, Z.F.; Pei, M.; Chen, Z.L.; Qu, L.J.; Gu, H.Y. Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem. Biophys. Res. Commun. 2006, 341, 1155–1163. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Sainz, M.B.; Grotewold, E.; Chandler, V.L. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 1997, 9, 611–625. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y. Research Progress of Kiwifruit in Tissue Culture. In Proceedings of the 2011 International Conference on Future Computer Science and Application (FCSA 2011), Hongkong, China, 18–19 June 2011; pp. 17–20. [Google Scholar]
- Akbas, F.; Isikalan, C.; Namli, S. Callus Induction and Plant Regeneration from Different Explants of Actinidia deliciosa. Appl. Biochem. Biotechnol. 2009, 158, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef]
- Lim, S.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Ha, S.H. A Radish Basic Helix-Loop-Helix Transcription Factor, RsTT8 Acts a Positive Regulator for Anthocyanin Biosynthesis. Front. Plant Sci. 2017, 8, 1917. [Google Scholar] [CrossRef]
- Hiscox, J.D.; Israelstam, G.F. Method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]
- Wang, A.; Li, R.; Ren, L.; Gao, X.; Zhang, Y.; Ma, Z.; Ma, D.; Luo, Y. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Palapol, Y.; Ketsa, S.; Kui, L.-W.; Ferguson, I.B.; Allan, A.C. A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta 2009, 229, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, B.; Qi, Y.; Liu, C.; Liu, Z.; Ren, X. Biochemical and functional characterization of AcUFGT3a, a galactosyltransferase involved in anthocyanin biosynthesis in the red-fleshed kiwifruit (Actinidia chinensis). Physiol. Plant 2018, 162, 409–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Atkinson, R.; Janssen, B. The choice of Agrobacterium strain for transformation of kiwifruit. Acta Hortic 2007, 753, 227–232. [Google Scholar] [CrossRef]
- Kumar, K.R.R.; Kirti, P.B. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol. Biochem. 2010, 48, 481–486. [Google Scholar] [CrossRef]










© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.; Man, Y.; Wang, Y. Light- and Temperature-Induced Expression of an R2R3-MYB Gene Regulates Anthocyanin Biosynthesis in Red-Fleshed Kiwifruit. Int. J. Mol. Sci. 2019, 20, 5228. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205228
Yu M, Man Y, Wang Y. Light- and Temperature-Induced Expression of an R2R3-MYB Gene Regulates Anthocyanin Biosynthesis in Red-Fleshed Kiwifruit. International Journal of Molecular Sciences. 2019; 20(20):5228. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205228
Chicago/Turabian StyleYu, Min, Yuping Man, and Yanchang Wang. 2019. "Light- and Temperature-Induced Expression of an R2R3-MYB Gene Regulates Anthocyanin Biosynthesis in Red-Fleshed Kiwifruit" International Journal of Molecular Sciences 20, no. 20: 5228. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205228