Pluripotent Cell Models for Gonadal Research
Abstract
:1. Introduction
2. Finding a Perfect Match: The Lack of Right Models for DSD Patients
2.1. Sertoli Cell Models
2.2. Granulosa Cell Models
2.3. Germ Cell Models
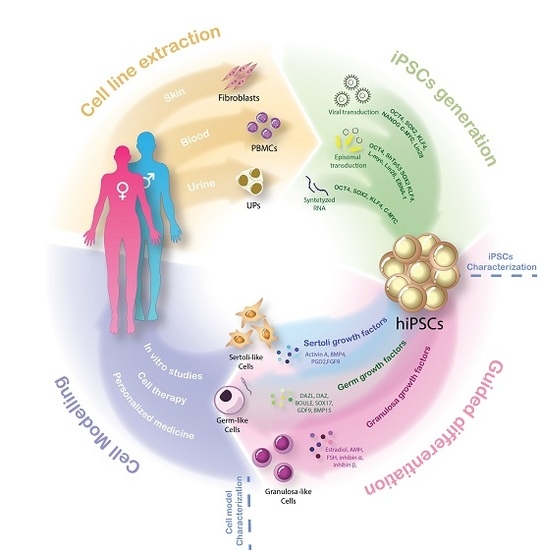
3. Unleashing the Power of Pluripotency: iPSCs and Guided Differentiation
4. Modeling the Complexity: Human iPSCs-Derived Models for DSD Research
4.1. hiPSCs and Somatic Gonadal Cell Models
4.2. hiPSCs and Germ Cell Models
5. Future of DSD Models
Limitations and Uncertainties
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franca, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.C.; Griswold, M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Zirkin, B.R.; Papadopoulos, V. Leydig cells: Formation, function, and regulation. Biol. Reprod. 2018, 99, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Lucas, T.F.; Nascimento, A.R.; Pisolato, R.; Pimenta, M.T.; Lazari, M.F.; Porto, C.S. Receptors and signaling pathways involved in proliferation and differentiation of Sertoli cells. Spermatogenesis 2014, 4, e28138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisostomo, L.; Alves, M.G.; Gorga, A.; Sousa, M.; Riera, M.F.; Galardo, M.N.; Meroni, S.B.; Oliveira, P.F. Molecular Mechanisms and Signaling Pathways Involved in the Nutritional Support of Spermatogenesis by Sertoli Cells. Methods Mol. Biol 2018, 1748, 129–155. [Google Scholar] [CrossRef]
- Kedem, A.; Yung, Y.; Yerushalmi, G.M.; Haas, J.; Maman, E.; Hanochi, M.; Hemi, R.; Orvieto, R.; Dor, J.; Hourvitz, A. Anti Mullerian Hormone (AMH) level and expression in mural and cumulus cells in relation to age. J. Ovarian Res. 2014, 7, 113. [Google Scholar] [CrossRef]
- Stocco, C. Aromatase expression in the ovary: Hormonal and molecular regulation. Steroids 2008, 73, 473–487. [Google Scholar] [CrossRef] [Green Version]
- Cocquet, J.; De Baere, E.; Gareil, M.; Pannetier, M.; Xia, X.; Fellous, M.; Veitia, R.A. Structure, evolution and expression of the FOXL2 transcription unit. Cytogenet. Genome Res. 2003, 101, 206–211. [Google Scholar] [CrossRef]
- Cocquet, J.; Pailhoux, E.; Jaubert, F.; Servel, N.; Xia, X.; Pannetier, M.; De Baere, E.; Messiaen, L.; Cotinot, C.; Fellous, M.; et al. Evolution and expression of FOXL2. J. Med. Genet. 2002, 39, 916–921. [Google Scholar] [CrossRef]
- Caburet, S.; Georges, A.; L’Hote, D.; Todeschini, A.L.; Benayoun, B.A.; Veitia, R.A. The transcription factor FOXL2: At the crossroads of ovarian physiology and pathology. Mol. Cell Endocrinol. 2012, 356, 55–64. [Google Scholar] [CrossRef]
- Pailhoux, E.; Vigier, B.; Chaffaux, S.; Servel, N.; Taourit, S.; Furet, J.P.; Fellous, M.; Grosclaude, F.; Cribiu, E.P.; Cotinot, C.; et al. A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat. Genet. 2001, 29, 453–458. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.C.; Klugmann, C.; Klasen, C.; Holter, N.I.; et al. Somatic Sex Reprogramming of Adult Ovaries to Testes by FOXL2 Ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maatouk, D.M.; Mork, L.; Chassot, A.A.; Chaboissier, M.C.; Capel, B. Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev. Biol. 2013, 383, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Megiorni, F.; Lin, L.; Mazzilli, M.C.; Gerrelli, D.; Majore, S.; Grammatico, P.; Achermann, J.C. Human RSPO1/R-spondin1 Is Expressed during Early Ovary Development and Augments beta-Catenin Signaling. PLoS ONE 2011, 6, e16366. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, J.V.; Kristensen, S.G.; Nielsen, M.E.; Humaidan, P.; Dal Canto, M.; Fadini, R.; Schmidt, K.T.; Ernst, E.; Yding Andersen, C. LH-receptor gene expression in human granulosa and cumulus cells from antral and preovulatory follicles. J. Clin. Endocrinol. Metab. 2012, 97, E1524–E1531. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Lane, M.; Thompson, J.G. Oocyte-secreted factors: Regulators of cumulus cell function and oocyte quality. Hum. Reprod. Update 2008, 14, 159–177. [Google Scholar] [CrossRef]
- Huang, Z.; Wells, D. The human oocyte and cumulus cells relationship: New insights from the cumulus cell transcriptome. Mol. Hum. Reprod. 2010, 16, 715–725. [Google Scholar] [CrossRef]
- Biason-Lauber, A.; Konrad, D.; Navratil, F.; Schoenle, E.J. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46,XX woman. N. Engl. J. Med. 2004, 351, 792–798. [Google Scholar] [CrossRef]
- Meyers-Wallen, V.N. Gonadal and Sex Differentiation Abnormalities of Dogs and Cats. Sex. Dev. 2012, 6, 46–60. [Google Scholar] [CrossRef]
- Parma, P.; Veyrunes, F.; Pailhoux, E. Sex Reversal in Non-Human Placental Mammals. Sex. Dev. 2016, 10, 326–344. [Google Scholar] [CrossRef]
- Lee, P.A.; Nordenström, A.; Houk, C.P.; Ahmed, S.F.; Auchus, R.; Baratz, A.; Baratz Dalke, K.; Liao, L.M.; Lin-Su, K.; Looijenga 3rd, L.H.J.; et al. Global disorders of sex development update since 2006: Perceptions, approach and care. Horm. Res. Paediat. 2016, 86, 158–180. [Google Scholar] [CrossRef]
- Eid, W.; Biason-Lauber, A. Why boys will be boys and girls will be girls: Human sex development and its defects. Birth Defects Res. C 2016, 108, 365–379. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Achermann, J.C.; Arlt, W.; Balen, A.; Conway, G.; Edwards, Z.; Elford, S.; Hughes, I.A.; Izatt, L.; Krone, N.; et al. Society for Endocrinology UK guidance on the initial evaluation of an infant or an adolescent with a suspected disorder of sex development (Revised 2015). Clin. Endocrinol. 2016, 84, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Edelsztein, N.Y.; Grinspon, R.P.; Schteingart, H.F.; Rey, R.A. Anti-Mullerian hormone as a marker of steroid and gonadotropin action in the testis of children and adolescents with disorders of the gonadal axis. Int. J. Pediatr. Endocrinol. 2016, 2016, 20. [Google Scholar] [CrossRef]
- Crisponi, L.; Deiana, M.; Loi, A.; Chiappe, F.; Uda, M.; Amati, P.; Bisceglia, L.; Zelante, L.; Nagaraja, R.; Porcu, S.; et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 2001, 27, 159–166. [Google Scholar] [CrossRef]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Megiorni, F.; De Bernardo, C.; Felici, A.; Marrocco, G.; Maggiulli, G.; Grammatico, B.; Remotti, D.; Saccucci, P.; Valentini, F.; et al. Syndromic true hermaphroditism due to an R-spondin1 (RSPO1) homozygous mutation. Hum. Mutat. 2008, 29, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Mather, J.P. Establishment and characterization of two distinct mouse testicular epithelial cell lines. Biol. Reprod. 1980, 23, 243–252. [Google Scholar] [CrossRef]
- Paquis-Flucklinger, V.; Michiels, J.F.; Vidal, F.; Alquier, C.; Pointis, G.; Bourdon, V.; Cuzin, F.; Rassoulzadegan, M. Expression in transgenic mice of the large T antigen of polyomavirus induces Sertoli cell tumours and allows the establishment of differentiated cell lines. Oncogene 1993, 8, 2087–2094. [Google Scholar]
- Larney, C.; Bailey, T.L.; Koopman, P. Switching on sex: Transcriptional regulation of the testis-determining gene Sry. Development 2014, 141, 2195–2205. [Google Scholar] [CrossRef]
- McClelland, K.; Bowles, J.; Koopman, P. Male sex determination: Insights into molecular mechanisms. Asian J. Androl. 2012, 14, 164–171. [Google Scholar] [CrossRef]
- Warr, N.; Bogani, D.; Siggers, P.; Brixey, R.; Tateossian, H.; Dopplapudi, A.; Wells, S.; Cheeseman, M.; Xia, Y.; Ostrer, H.; et al. Minor abnormalities of testis development in mice lacking the gene encoding the MAPK signalling component, MAP3K1. PLoS ONE 2011, 6, e19572. [Google Scholar] [CrossRef] [PubMed]
- Chui, K.; Trivedi, A.; Cheng, C.Y.; Cherbavaz, D.B.; Dazin, P.F.; Ai, L.T.H.; Mitchell, J.B.; Rabinovich, G.A.; Noble-Haeusslein, L.J.; John, C.M. Characterization and Functionality of Proliferative Human Sertoli Cells. Cell Transplant. 2011, 20, 619–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, E.A.; Barten-van Rijbroek, A.D.; Kal, H.B.; Sadri-Ardekani, H.; Mizrak, S.C.; van Pelt, A.M.; de Rooij, D.G. Proliferative activity in vitro and DNA repair indicate that adult mouse and human Sertoli cells are not terminally differentiated, quiescent cells. Biol. Reprod. 2009, 80, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hai, Y.; Yao, C.; Chen, Z.; Hou, J.; Li, Z.; He, Z. Long-term culture and significant expansion of human Sertoli cells whilst maintaining stable global phenotype and AKT and SMAD1/5 activation. Cell Commun. Signal. 2015, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Andrews, P.W.; Damjanov, I.; Simon, D.; Banting, G.S.; Carlin, C.; Dracopoli, N.C.; Fogh, J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 1984, 50, 147–162. [Google Scholar] [PubMed]
- De Santa Barbara, P.; Bonneaud, N.; Boizet, B.; Desclozeaux, M.; Moniot, B.; Sudbeck, P.; Scherer, G.; Poulat, F.; Berta, P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol. Cell Biol. 1998, 18, 6653–6665. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Gutierrez, D.; Eid, W.; Biason-Lauber, A. A Human Gonadal Cell Model From Induced Pluripotent Stem Cells. Front. Genet. 2018, 9, 498. [Google Scholar] [CrossRef]
- Alexopoulos, E.; Shahid, J.; Ongley, H.Z.; Richardson, M.C. Luteinized human granulosa cells are associated with endogenous basement membrane-like components in culture. Mol. Hum. Reprod. 2000, 6, 324–330. [Google Scholar] [CrossRef]
- Bruckova, L.; Soukup, T.; Visek, B.; Moos, J.; Moosova, M.; Pavelkova, J.; Rezabek, K.; Kucerova, L.; Micuda, S.; Brcakova, E.; et al. Proliferative potential and phenotypic analysis of long-term cultivated human granulosa cells initiated by addition of follicular fluid. J. Assist. Reprod. Genet. 2011, 28, 939–950. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.L.; Kendall, J.Z.; Dandekar, P.V.; Quigley, M.M.; Schmidt, K.L. Characterization of long-term monolayer cultures of human granulosa cells from follicles of different size and exposed in vivo to clomiphene citrate and hCG. J. Reprod. Fertil. 1984, 71, 279–287. [Google Scholar] [CrossRef]
- Lie, B.L.; Leung, E.; Leung, P.C.; Auersperg, N. Long-term growth and steroidogenic potential of human granulosa-lutein cells immortalized with SV40 large T antigen. Mol. Cell Endocrinol. 1996, 120, 169–176. [Google Scholar] [CrossRef]
- Bruckova, L.; Soukup, T.; Moos, J.; Moosova, M.; Pavelkova, J.; Rezabek, K.; Visek, B.; Mokry, J. The cultivation of human granulosa cells. Acta Med. (Hradec Kral.) 2008, 51, 165–172. [Google Scholar] [CrossRef]
- Nishi, Y.; Yanase, T.; Mu, Y.; Oba, K.; Ichino, I.; Saito, M.; Nomura, M.; Mukasa, C.; Okabe, T.; Goto, K.; et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 2001, 142, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg-Bakker, C.A.; Hagemeijer, A.; Franken-Postma, E.M.; Smit, V.T.; Kuppen, P.J.; van Ravenswaay Claasen, H.H.; Cornelisse, C.J.; Schrier, P.I. Establishment and characterization of 7 ovarian carcinoma cell lines and one granulosa tumor cell line: Growth features and cytogenetics. Int. J. Cancer 1993, 53, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Klausen, C.; Leung, P.C. Overexpression of wild-type but not C134W mutant FOXL2 enhances GnRH-induced cell apoptosis by increasing GnRH receptor expression in human granulosa cell tumors. PLoS ONE 2013, 8, e55099. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.; Butzow, R.; Andersson, N.; Alexiadis, M.; Unkila-Kallio, L.; Heikinheimo, M.; Fuller, P.J.; Anttonen, M. The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Mod. Pathol. 2010, 23, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, K.; Dantes, A.; Schere-Levy, C.; Barash, A.; Yoshida, Y.; Kotsuji, F.; Vlodavsky, I.; Amsterdam, A. Induction of Ad4BP/SF-1, steroidogenic acute regulatory protein, and cytochrome P450scc enzyme system expression in newly established human granulosa cell lines. Endocrinology 1998, 139, 4679–4687. [Google Scholar] [CrossRef]
- Rainey, W.H.; Sawetawan, C.; Shay, J.W.; Michael, M.D.; Mathis, J.M.; Kutteh, W.; Byrd, W.; Carr, B.R. Transformation of Human Granulosa-Cells with the E6 and E7 Regions of Human Papillomavirus. J. Clin. Endocr. Metab. 1994, 78, 705–710. [Google Scholar] [CrossRef]
- Iwase, A.; Kiyono, T.; Takikawa, S.; Goto, M.; Nakamura, T.; Nagatomo, Y.; Nakahara, T.; Kotani, T.; Kobayashi, H.; Kondo, M.; et al. Establishment of a human nonluteinized granulosa cell line that transitions from the gonadotropin-independent to the gonadotropin-dependent status. Endocrinology 2012, 153, 2851–2860. [Google Scholar] [CrossRef]
- Di Pasquale, E.; Rossetti, R.; Marozzi, A.; Bodega, B.; Borgato, S.; Cavallo, L.; Einaudi, S.; Radetti, G.; Russo, G.; Sacco, M.; et al. Identification of new variants of human BMP15 gene in a large cohort of women with premature ovarian failure. J. Clin. Endocrinol. Metab. 2006, 91, 1976–1979. [Google Scholar] [CrossRef]
- Galloway, S.M.; McNatty, K.P.; Cambridge, L.M.; Laitinen, M.P.; Juengel, J.L.; Jokiranta, T.S.; McLaren, R.J.; Luiro, K.; Dodds, K.G.; Montgomery, G.W.; et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat. Genet. 2000, 25, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wang, P.; DeMayo, J.; DeMayo, F.J.; Elvin, J.A.; Carino, C.; Prasad, S.V.; Skinner, S.S.; Dunbar, B.S.; Dube, J.L.; et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol. Endocrinol. 2001, 15, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Lesch, B.J.; Page, D.C. Genetics of germ cell development. Nat. Rev. Genet. 2012, 13, 781–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irie, N.; Tang, W.W.; Azim Surani, M. Germ cell specification and pluripotency in mammals: A perspective from early embryogenesis. Reprod. Med. Biol. 2014, 13, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Weinberger, L.; Tang, W.W.C.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 Is a Critical Specifier of Human Primordial Germ Cell Fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar] [CrossRef]
- Geijsen, N.; Horoschak, M.; Kim, K.; Gribnau, J.; Eggan, K.; Daley, G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004, 427, 148–154. [Google Scholar] [CrossRef]
- Ko, K.; Huebner, K.; Mueller-Keuker, J.; Schoeler, H.R. In vitro derivation of germ cells from embryonic stem cells. Front. Biosci. 2010, 15, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, E.; Forabosco, A.; Schlessinger, D. Germ cell formation from embryonic stem cells and the use of somatic cell nuclei in oocytes. Ann. N. Y. Acad. Sci. 2011, 1221, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Toyooka, Y.; Tsunekawa, N.; Akasu, R.; Noce, T. Embryonic stem cells can form germ cells in vitro. Proc. Natl. Acad Sci. USA 2003, 100, 11457–11462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.B.; Meng, Q.G.; Li, N. In Vitro Derivation of Germ Cells From Embryonic Stem Cells in Mammals. Mol. Reprod. Dev. 2010, 77, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Gurdon, J.B. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 1962, 10, 622–640. [Google Scholar] [PubMed]
- Tada, M.; Takahama, Y.; Abe, K.; Nakatsuji, N.; Tada, T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001, 11, 1553–1558. [Google Scholar] [CrossRef] [Green Version]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H. Viable offspring derived from fetal and adult mammalian cells. Nature 1997, 385, 810–813. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Schneuwly, S.; Klemenz, R.; Gehring, W.J. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature 1987, 325, 816–818. [Google Scholar] [CrossRef]
- Yamanaka, S.; Blau, H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010, 465, 704–712. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Smith, A.G.; Heath, J.K.; Donaldson, D.D.; Wong, G.G.; Moreau, J.; Stahl, M.; Rogers, D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 1988, 336, 688–690. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Puri, M.C.; Nagy, A. Concise review: Embryonic stem cells versus induced pluripotent stem cells: The game is on. Stem Cells 2012, 30, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Cassady, J.P.; Jaenisch, R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008, 2, 10–12. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef]
- Yu, J.Y.; Hu, K.J.; Smuga-Otto, K.; Tian, S.L.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [Green Version]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Jpn. Ser. B 2009, 85, 348–362. [Google Scholar] [CrossRef] [Green Version]
- Bang, J.S.; Choi, N.Y.; Lee, M.; Ko, K.; Lee, H.J.; Park, Y.S.; Jeong, D.; Chung, H.M.; Ko, K. Optimization of episomal reprogramming for generation of human induced pluripotent stem cells from fibroblasts. Anim. Cells Syst. 2018, 22, 132–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, P.K.; Rossi, D.J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013, 8, 568–582. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wang, Q. Efficient Generation of Non-Integration and Feeder-Free Induced Pluripotent Stem Cells from Human Peripheral Blood Cells by Sendai Virus. Cell Physiol. Biochem. 2018, 50, 1318–1331. [Google Scholar] [CrossRef] [PubMed]
- Nefzger, C.M.; Rossello, F.J.; Chen, J.; Liu, X.D.; Knaupp, A.S.; Firas, J.; Paynter, J.M.; Pflueger, J.; Buckberry, S.; Lim, S.M.; et al. Cell Type of Origin Dictates the Route to Pluripotency. Cell Rep. 2017, 21, 2649–2660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, A. DNA methylation patterns and epigenetic memory. Gene Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, Z.; Wilson, K.D.; Wu, Y.; Hu, S.J.; Quertermous, T.; Wu, J.C. Persistent Donor Cell Gene Expression among Human Induced Pluripotent Stem Cells Contributes to Differences with Human Embryonic Stem Cells. PLoS ONE 2010, 5, e8975. [Google Scholar] [CrossRef] [PubMed]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerias, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012, 22, 168–177. [Google Scholar] [CrossRef]
- Spyrou, J.; Gardner, D.K.; Harvey, A.J. Metabolism Is a Key Regulator of Induced Pluripotent Stem Cell Reprogramming. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef]
- Setthawong, P.; Phakdeedindan, P.; Tiptanavattana, N.; Rungarunlert, S.; Techakumphu, M.; Tharasanit, T. Generation of porcine induced-pluripotent stem cells from Sertoli cells. Theriogenology 2019, 127, 32–40. [Google Scholar] [CrossRef]
- Loh, Y.H.; Hartung, O.; Li, H.; Guo, C.G.; Sahalie, J.M.; Manos, P.D.; Urbach, A.; Heffner, G.C.; Grskovic, M.; Vigneault, F.; et al. Reprogramming of T Cells from Human Peripheral Blood. Cell Stem Cell 2010, 7, 15–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Babot, G.; Balyura, M.; Hadjidemetriou, I.; Ajodha, S.J.; Taylor, D.R.; Ghataore, L.; Taylor, N.F.; Schubert, U.; Ziegler, C.G.; Storr, H.L.; et al. Modeling Congenital Adrenal Hyperplasia and Testing Interventions for Adrenal Insufficiency Using Donor-Specific Reprogrammed Cells. Cell Rep. 2018, 22, 1236–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.; Benda, C.; Duzinger, S.; Huang, Y.H.; Li, X.Y.; Li, Y.H.; Guo, X.P.; Cao, G.K.; Chen, S.; Hao, L.L.; et al. Generation of Induced Pluripotent Stem Cells from Urine. J. Am. Soc. Nephrol. 2011, 22, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Marti, M.; Mulero, L.; Pardo, C.; Morera, C.; Carrio, M.; Laricchia-Robbio, L.; Esteban, C.R.; Izpisua Belmonte, J.C. Characterization of pluripotent stem cells. Nat. Protoc. 2013, 8, 223–253. [Google Scholar] [CrossRef]
- Sekine, K.; Takebe, T.; Suzuki, Y.; Kamiya, A.; Nakauchi, H.; Taniguchi, H. Highly Efficient Generation of Definitive Endoderm Lineage from Human Induced Pluripotent Stem Cells. Transpl. Proc. 2012, 44, 1127–1129. [Google Scholar] [CrossRef]
- Terryn, J.; Tricot, T.; Gajjar, M.; Verfaillie, C. Recent advances in lineage differentiation from stem cells: Hurdles and opportunities? F1000Res 2018, 7, 220. [Google Scholar] [CrossRef]
- Bucay, N.; Yebra, M.; Cirulli, V.; Afrikanova, I.; Kaido, T.; Hayek, A.; Montgomery, A.M.P. A Novel Approach for the Derivation of Putative Primordial Germ Cells and Sertoli Cells from Human Embryonic Stem Cells. Stem Cells 2009, 27, 68–77. [Google Scholar] [CrossRef]
- Shlush, E.; Maghen, L.; Swanson, S.; Kenigsberg, S.; Moskovtsev, S.; Barretto, T.; Gauthier-Fisher, A.; Librach, C.L. In vitro generation of Sertoli-like and haploid spermatid-like cells from human umbilical cord perivascular cells. Stem Cell Res. Ther. 2017, 8, 37. [Google Scholar] [CrossRef]
- Anchan, R.; Gerami-Naini, B.; Lindsey, J.S.; Ho, J.W.K.; Kiezun, A.; Lipskind, S.; Ng, N.; LiCausi, J.A.; Kim, C.S.; Brezina, P.; et al. Efficient Differentiation of Steroidogenic and Germ-Like Cells from Epigenetically-Related iPSCs Derived from Ovarian Granulosa Cells. PLoS ONE 2015, 10, e0119275. [Google Scholar] [CrossRef]
- Liu, T.; Li, Q.; Wang, S.; Chen, C.; Zheng, J. Transplantation of ovarian granulosalike cells derived from human induced pluripotent stem cells for the treatment of murine premature ovarian failure. Mol. Med. Rep. 2016, 13, 5053–5058. [Google Scholar] [CrossRef]
- Lipskind, S.; Lindsey, J.S.; Gerami-Naini, B.; Eaton, J.L.; O’Connell, D.; Kiezun, A.; Ho, J.W.K.; Ng, N.; Parasar, P.; Ng, M.; et al. An Embryonic and Induced Pluripotent Stem Cell Model for Ovarian Granulosa Cell Development and Steroidogenesis. Reprod. Sci. 2018, 25, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Kee, K.; Angeles, V.T.; Flores, M.; Nguyen, H.N.; Pera, R.A.R. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 2009, 462, U222–U295. [Google Scholar] [CrossRef] [PubMed]
- Eguizabal, C.; Montserrat, N.; Vassena, R.; Barragan, M.; Garreta, E.; Garcia-Quevedo, L.; Vidal, F.; Giorgetti, A.; Veiga, A.; Belmonte, J.C.I. Complete Meiosis from Human Induced Pluripotent Stem Cells. Stem Cells 2011, 29, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Medrano, J.V.; Ramathal, C.; Nguyen, H.N.; Simon, C.; Pera, R.A.R. Divergent RNA-Binding Proteins, DAZL and VASA, Induce Meiotic Progression in Human Germ Cells Derived In Vitro. Stem Cells 2012, 30, 441–451. [Google Scholar] [CrossRef]
- Yang, S.; Ding, S.F.; He, S.W.; He, L.X.; Gao, K.F.; Peng, S.P.; Shuai, C.J. Differentiation of primordial germ cells from premature ovarian insufficiency-derived induced pluripotent stem cells. Stem Cell Res. Ther. 2019, 10, 156. [Google Scholar] [CrossRef]
- Panula, S.; Medrano, J.V.; Kee, K.; Bergstrom, R.; Ha, N.N.; Byers, B.; Wilson, K.D.; Wu, J.C.; Simon, C.; Hovatta, O.; et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 752–762. [Google Scholar] [CrossRef]
- Easley, C.A.; Phillips, B.T.; McGuire, M.M.; Barringer, J.M.; Valli, H.; Hermann, B.P.; Simerly, C.R.; Rajkovic, A.; Miki, T.; Orwig, K.E.; et al. Direct Differentiation of Human Pluripotent Stem Cells into Haploid Spermatogenic Cells. Cell Rep. 2012, 2, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.J.; Xiong, J.; Ye, M.; Qin, X.S.; Li, L.; Cheng, S.F.; Luo, M.Y.; Peng, J.; Dong, J.; Tang, F.C.; et al. In vitro differentiation of human embryonic stem cells into ovarian follicle-like cells. Nat. Commun. 2017, 8, 15680. [Google Scholar] [CrossRef]
- Buganim, Y.; Itskovich, E.; Hu, Y.C.; Cheng, A.W.; Ganz, K.; Sarkar, S.; Fu, D.D.; Welstead, G.G.; Page, D.C.; Jaenisch, R. Direct Reprogramming of Fibroblasts into Embryonic Sertoli-like Cells by Defined Factors. Cell Stem Cell 2012, 11, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Evseenko, D.; Zhu, Y.H.; Schenke-Layland, K.; Kuo, J.; Latour, B.; Ge, S.D.; Scholes, J.; Dravid, G.; Li, X.M.; MacLellan, W.R.; et al. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 13742–13747. [Google Scholar] [CrossRef] [Green Version]
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.J.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.E.; Korving, J.P.W.F.M.; Hogan, B.L.M. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Gene Dev. 1999, 13, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Kan, M.; McKeehan, W.L.; de Crombrugghe, B. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 2000, 97, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kobayashi, A.; Sekido, R.; DiNapoli, L.; Brennan, J.; Chaboissier, M.C.; Poulat, F.; Behringer, R.R.; Lovell-Badge, R.; Capel, B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006, 4, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Moniot, B.; Declosmenil, F.; Barrionuevo, F.; Scherer, G.; Aritake, K.; Malki, S.; Marzi, L.; Cohen-Solal, A.; Georg, I.; Klattig, J.; et al. The PGD2 pathway, independently of FGF9, amplifies SOX9 activity in Sertoli cells during male sexual differentiation. Development 2009, 136, 1813–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anamthathmakula, P.; Miryala, C.S.J.; Moreci, R.S.; Kyathanahalli, C.; Hassan, S.S.; Condon, J.C.; Jeyasuria, P. Steroidogenic Factor 1 (Nr5a1) is Required for Sertoli Cell Survival Post Sex Determination. Sci. Rep. 2019, 9, 4452. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Stanton, P.G.; Chen, J.L.; Olcorn, J.S.; Haverfield, J.T.; Qian, H.; Walton, K.L.; Gregorevic, P.; Harrison, C.A. Activin signaling regulates Sertoli cell differentiation and function. Endocrinology 2012, 153, 6065–6077. [Google Scholar] [CrossRef]
- Kang, Y.; Cheng, M.J.; Xu, C.J. Secretion of oestrogen from murine-induced pluripotent stem cells co-cultured with ovarian granulosa cells in vitro. Cell Biol Int 2011, 35, 871–874. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Wu, Z.; Tan, X.J.; Liu, F.Y.; Huang, X.H.; Fang, X.L. Differentiation of rat iPS cells and ES cells into granulosa cell-like cells in vitro. Acta. Bioch. Bioph. Sin. 2013, 45, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Hubner, K.; Fuhrmann, G.; Christenson, L.K.; Kehler, J.; Reinbold, R.; De La Fuente, R.; Wood, J.; Strauss, J.F.; Boiani, M.; Scholer, H.R. Derivation of oocytes from mouse embryonic stem cells. Science 2003, 300, 1251–1256. [Google Scholar] [CrossRef]
- Nakaki, F.; Hayashi, K.; Ohta, H.; Kurimoto, K.; Yabuta, Y.; Saitou, M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature 2013, 501, 222. [Google Scholar] [CrossRef]
- Tang, W.W.C.; Dietmann, S.; Irie, N.; Leitch, H.G.; Floros, V.I.; Bradshaw, C.R.; Hackett, J.A.; Chinnery, P.F.; Surani, M.A. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 2015, 161, 1453–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Kawaguchi, T.; Durcova-Hills, G.; Imai, H. Generation of germ cells from pluripotent stem cells in mammals. Reprod. Med. Biol. 2018, 17, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Eirin-Lopez, J.M.; Ausio, J. Boule and the Evolutionary Origin of Metazoan Gametogenesis: A Grandpa’s Tale. Int. J. Evol. Biol. 2011, 2011, 972457. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.Y.; Moore, F.L.; Pera, R.A.R. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. USA 2001, 98, 7414–7419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panula, S.; Reda, A.; Stukenborg, J.B.; Ramathal, C.; Sukhwani, M.; Albalushi, H.; Edsgard, D.; Nakamura, M.; Soder, O.; Orwig, K.E.; et al. Over Expression of NANOS3 and DAZL in Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0165268. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Ye, S.C.; Liang, D.L.; Wang, P.X.; Fu, J.; Ma, Q.; Kong, R.J.; Shi, L.H.; Gong, X.P.; Chen, W.; et al. In Vitro Modeling of Human Germ Cell Development Using Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 509–523. [Google Scholar] [CrossRef] [Green Version]
- Kee, K.; Gonsalves, J.M.; Clark, A.T.; Pera, R.A.R. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006, 15, 831–837. [Google Scholar] [CrossRef]
- Hurrell, T.; Segeritz, C.P.; Vallier, L.; Lilley, K.S.; Cromarty, A.D. A proteomic time course through the differentiation of human induced pluripotent stem cells into hepatocyte-like cells. Sci. Rep. 2019, 9, 3270. [Google Scholar] [CrossRef]
- Ben Jehuda, R.; Shemer, Y.; Binah, O. Genome Editing in Induced Pluripotent Stem Cells using CRISPR/Cas9. Stem Cell Rev. Rep. 2018, 14, 323–336. [Google Scholar] [CrossRef]
- Chang, C.Y.; Ting, H.C.; Su, H.L.; Jeng, J.R. Combining Induced Pluripotent Stem Cells and Genome Editing Technologies for Clinical Applications. Cell Transplant. 2018, 27, 379–392. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, E.; Chen, T. A matter of life and death: Self-renewal in stem cells. Embo. Rep. 2013, 14, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Pennarossa, G.; Santoro, R.; Manzoni, E.F.M.; Pesce, M.; Gandolfi, F.; Brevini, T.A.L. Epigenetic Erasing and Pancreatic Differentiation of Dermal Fibroblasts into Insulin-Producing Cells are Boosted by the Use of Low-Stiffness Substrate. Stem Cell Rev. Rep. 2018, 14, 398–411. [Google Scholar] [CrossRef] [PubMed]
- McCauley, H.A.; Wells, J.M. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Development 2017, 144, 958–962. [Google Scholar] [CrossRef] [PubMed]
- Laronda, M.M.; Burdette, J.E.; Kim, J.J.; Woodruff, T.K. Recreating the female reproductive tract in vitro using iPSC technology in a linked microfluidics environment. Stem Cell Res. Ther. 2013, 4, S13. [Google Scholar] [CrossRef] [PubMed]
- Meroni, S.B.; Galardo, M.N.; Rindone, G.; Gorga, A.; Riera, M.F.; Cigorraga, S.B. Molecular Mechanisms and Signaling Pathways Involved in Sertoli Cell Proliferation. Front. Endocrinol. 2019, 10, 224. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, J.Y.; Romero, M.M.; Smith, K.N.; Shea, L.D.; Woodruff, T.K. In vitro follicle growth supports human oocyte meiotic maturation. Sci Rep. 2015, 5, 17323. [Google Scholar] [CrossRef]
- Quaas, A.; Dokras, A. Diagnosis and treatment of unexplained infertility. Rev. Obs. Gynecol. 2008, 1, 69–76. [Google Scholar]



| Cell Type | Cell Model | Differentiation Mechanism | Remarks | Reference |
|---|---|---|---|---|
| Sertoli cell | SLC | Unguided differentiation in co-culture with PGCs | Possible paracrine action of PGCs on differentiation of SLCs | Bucay et al., 2009 [97] |
| SLC | 5-step differentiation protocol, including RA, LIF, GDNF, putrescine, testosterone and FSH | BMP4 secretion by undifferentiated cells | Shlush et al., 2017 [98] | |
| SLC | Addition of BMP4, PGDS, bFGF, FGF9 and Activin-A to growth medium | Transcriptomic landscape resemble differentiating Sertoli cells | Rodríguez Gutiérrez et al., 2018 [37] | |
| Granulosa cell | Ovarian steroidogenic cells | Redifferentiation of iPSCs derived from GCs into homotypic ovarian steroidogenic cells | Cellular origin have strong effect on final differentiation faith | Anchan et al., 2015 [99] |
| GLC | Addition of all-trans-retinoic acid, estradiol, AMH, FSH, Inhibin α, Inhibin β and TGF –β to growth medium | GLCs can rescue ovarian failure when transplanted in POF mice. | Liu et al., 2016 [100] | |
| GLC | Differentiation from human amniocytes without intervention of additional growth factors. | GLC able to synthetize E2 | Lipskind et al., 2018 [101] | |
| Primordial germ cell | hPGCLCs | Differentiation induction by recombinant BMPs | BMPs induce differentiation of germ cells from hES cells | Kee et al., 2009 [102] |
| Haploid Gamete-like Cells | Two step protocol: Culture in bFGF-depleted ES cell media followed by addition of RA | Epigenetic memory of the reprogrammed somatic cells | Eguizabal et al.,2011 [103] | |
| Meiotic GCs | Overexpression of VASA and/or DAZL following differentiation on matrigel-coated plates | Differentiation of germ cells is dependent on post-translational regulation by RNA-binding proteins like VASA | Medrano et al., 2011 [104] | |
| hPGCLCs | Addition of BMP2 or BMP4, LIF, SCF, EGF and ROCK inhibitor to growth medium | SOX17 is the key regulator of hPGCLC specification | Irie et al., 2015 [55] | |
| hPGCLCs | (1) in vitro: same protocol as Irie et al., 2015 (2) in vivo: xenotransplantation into NOD/SCID mice | hPGCLCs had the potential for meiotic progression in vitro | Yang et al., 2019 [105] | |
| Spermatid | SpLC | Differentiation induction by BMPs followed by overexpression of DAZ, DAZL, and BOULE | hiPSC lines can differentiate to haploid cells with characteristic staining of ACROSIN for spermatid. | Panula et al., 2011 [106] |
| SSC | Direct differentiation using mouse spermatogonial stem cell (SSC) medium | hPSCs differentiate into spermatogonia, spermatocytes and haploid spermatids | Easley et al., 2012 [107] | |
| Oocyte | FLCs | Overexpression of DAZL and BOULE with recombinant human GDF9 and BMP15 | GDF9 and BMP15 induce ovarian follicle formation in hESCs | Jung et al.,2017 [108] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Gutiérrez, D.; Biason-Lauber, A. Pluripotent Cell Models for Gonadal Research. Int. J. Mol. Sci. 2019, 20, 5495. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215495
Rodríguez Gutiérrez D, Biason-Lauber A. Pluripotent Cell Models for Gonadal Research. International Journal of Molecular Sciences. 2019; 20(21):5495. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215495
Chicago/Turabian StyleRodríguez Gutiérrez, Daniel, and Anna Biason-Lauber. 2019. "Pluripotent Cell Models for Gonadal Research" International Journal of Molecular Sciences 20, no. 21: 5495. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215495