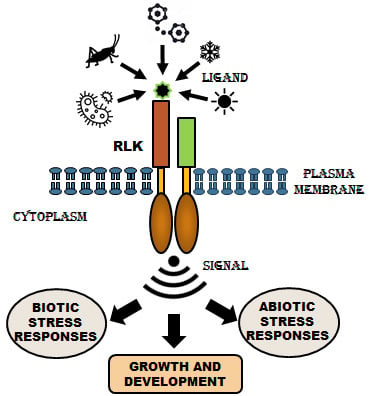
Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery
Abstract
:1. Introduction
2. Classification of Arabidopsis RLKs
3. Signaling Mechanism of RLKs
4. Functions of RLKs in the Regulation of Plant Growth and Development
4.1. Regulation in Anther and Ovule Development
4.2. Pollen-Pistil Interactions
4.3. Role in Embryo Development
4.4. Organ Development
4.5. Vascular Tissue Development
4.6. Regulation of Organ Abscission
4.7. Modulation of Phytohormone Signaling
5. RLKs in the Regulation of Plant Biotic Interactions
5.1. RLKs in Pathogen Triggered Immunity
5.2. RLKs in Effector Triggered Immunity
5.3. CRKs in Defense and Hypersensitive Responses
6. RLKs in the Regulation of Plant Abiotic Stresses
7. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAK1 | BRI1-associated receptor kinase 1 |
| DAMP | Damage-associated molecular patterns |
| LRRs | Leucine-rich repeats |
| LysM | Lysin motif |
| MAMP | Microbe-associated molecular patterns |
| PAMP | Pathogen-associated molecular patterns |
| RLCK | Receptor-like cytoplasmic kinase |
| RLK | Receptor-like kinase |
| RLP | Receptor-like protein |
| SERK | Somatic embryogenesis receptor kinase |
References
- Lehti-Shiu, M.D.; Zou, C.; Shiu, S.-H. Origin, Diversity, Expansion history, and functional evolution of the plant receptor-like kinase/pelle family. In Receptor-Like Kinases in Plants: From Development to Defense; Tax, F., Kemmerling, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–22. [Google Scholar]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cock, J.M.; Vanoosthuyse, V.; Gaude, T. Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 2002, 14, 230–236. [Google Scholar] [CrossRef]
- Roux, M.; Zipfel, C. Receptor kinase interactions: Complexity of signalling. In Receptor-Like Kinases in Plants: From Development to Defense; Tax, F., Kemmerling, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 145–172. [Google Scholar]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, J.; Shan, L.; Meng, X. Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 2018, 131, jcs209353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [Green Version]
- Turra, D.; Segorbe, D.; Di Pietro, A. Protein kinases in plant-pathogenic fungi: Conserved regulators of infection. Annu. Rev. Phytopathol. 2014, 52, 267–288. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [Green Version]
- Oh, M.H.; Wang, X.; Kota, U.; Goshe, M.B.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Madsen, E.B.; Antolin-Llovera, M.; Grossmann, C.; Ye, J.; Vieweg, S.; Broghammer, A.; Krusell, L.; Radutoiu, S.; Jensen, O.N.; Stougaard, J.; et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011, 65, 404–417. [Google Scholar] [CrossRef]
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006, 16, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 1999, 11, 1925–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, C.; Russinova, E.; Hecht, V.; Baaijens, E.; de Vries, S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 2005, 17, 3337–3349. [Google Scholar] [CrossRef] [Green Version]
- Jinn, T.L.; Stone, J.M.; Walker, J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000, 14, 108–117. [Google Scholar]
- Gomez-Gomez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Verica, J.A.; He, Z.H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Baumberger, N.; Ringli, C.; Keller, B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001, 15, 1128–1139. [Google Scholar] [CrossRef] [Green Version]
- Engelsdorf, T.; Hamann, T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann. Bot. 2014, 114, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Huck, N.; Moore, J.M.; Federer, M.; Grossniklaus, U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 2003, 130, 2149–2159. [Google Scholar] [CrossRef] [Green Version]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [Green Version]
- Gifford, M.L.; Dean, S.; Ingram, G.C. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 2003, 130, 4249–4258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czyzewicz, N.; Nikonorova, N.; Meyer, M.R.; Sandal, P.; Shah, S.; Vu, L.D.; Gevaert, K.; Rao, A.G.; De Smet, I. The growing story of (ARABIDOPSIS) CRINKLY 4. J. Exp. Bot. 2016, 67, 4835–4847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, B.R.; Raina, S.; Maqbool, S.B.; Jagadeeswaran, G.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007, 50, 488–499. [Google Scholar] [CrossRef]
- Idanheimo, N.; Gauthier, A.; Salojarvi, J.; Siligato, R.; Brosche, M.; Kollist, H.; Mahonen, A.P.; Kangasjarvi, J.; Wrzaczek, M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Willmann, R.; Lajunen, H.M.; Erbs, G.; Newman, M.A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zafian, P.; Choudhary, M.; Lawton, M. The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 2598–2602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.W.; Yang, S.H.; Shin, K.H.; Lee, S.C.; Kim, S.H. The AtLRK10L1.2, Arabidopsis ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 2015, 34, 447–455. [Google Scholar] [CrossRef]
- Bellande, K.; Bono, J.J.; Savelli, B.; Jamet, E.; Canut, H. Plant Lectins and lectin receptor-like kinases: How do they sense the outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef]
- Dwyer, K.G.; Kandasamy, M.K.; Mahosky, D.I.; Acciai, J.; Kudish, B.I.; Miller, J.E.; Nasrallah, M.E.; Nasrallah, J.B. A superfamily of S locus-related sequences in Arabidopsis: Diverse structures and expression patterns. Plant Cell 1994, 6, 1829–1843. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.A.; Lindner, H.; Jones, D.S.; Grossniklaus, U. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 2015, 16, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Dunne, A.; O’Neill, L.A. The interleukin-1 receptor/Toll-like receptor superfamily: Signal transduction during inflammation and host defense. Sci. STKE 2003, 2003, re3. [Google Scholar] [CrossRef] [PubMed]
- Ten Hove, C.A.; Bochdanovits, Z.; Jansweijer, V.M.; Koning, F.G.; Berke, L.; Sanchez-Perez, G.F.; Scheres, B.; Heidstra, R. Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 2011, 76, 69–83. [Google Scholar] [CrossRef] [Green Version]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Wang, H.; Chevalier, D.; Larue, C.; Ki Cho, S.; Walker, J.C. The protein phosphatases and protein kinases of Arabidopsis thaliana. Arab. Book 2007, 5, e0106. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. Legume lectins—A large family of homologous proteins. FASEB J. 1990, 4, 3198–3208. [Google Scholar] [CrossRef]
- Cambi, A.; Koopman, M.; Figdor, C.G. How C-type lectins detect pathogens. Cell Microbiol. 2005, 7, 481–488. [Google Scholar] [CrossRef]
- Herve, C.; Serres, J.; Dabos, P.; Canut, H.; Barre, A.; Rouge, P.; Lescure, B. Characterization of the Arabidopsis lecRK-a genes: Members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol. Biol. 1999, 39, 671–682. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef] [Green Version]
- Cassab, G.I. Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 281–309. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.N.; Lee, Y.S.; Cho, L.H.; Jeong, H.J.; An, G.; Jung, K.H. Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 2015, 241, 603–613. [Google Scholar] [CrossRef]
- Schallus, T.; Jaeckh, C.; Feher, K.; Palma, A.S.; Liu, Y.; Simpson, J.C.; Mackeen, M.; Stier, G.; Gibson, T.J.; Feizi, T.; et al. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 2008, 19, 3404–3414. [Google Scholar] [CrossRef] [Green Version]
- Nasrallah, J.B.; Yu, S.M.; Nasrallah, M.E. Self-incompatibility genes of Brassica oleracea: Expression, isolation, and structure. Proc. Natl. Acad. Sci. USA 1988, 85, 5551–5555. [Google Scholar] [CrossRef] [Green Version]
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Vaattovaara, A.; Brandt, B.; Rajaraman, S.; Safronov, O.; Veidenberg, A.; Luklová, M.; Kangasjärvi, J.; Löytynoja, A.; Hothorn, M.; Salojärvi, J.; et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019, 2, 56. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-like kinase and lysm receptor-like protein families: An update on phylogeny and functional characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, S.R.; Pandey, S. Phosphorylation-dependent regulation of G-protein cycle during nodule formation in soybean. Plant Cell 2015, 27, 3260–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef] [Green Version]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [Green Version]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef] [Green Version]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.D.; Li, J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012, 8, e1002452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotochaud, A.E.; Hao, T.; Wu, G.; Yang, Z.; Clark, S.E. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 1999, 11, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Becraft, P.W. Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 2002, 18, 163–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda-Sicilia, M.N.; Trusov, Y.; Maruta, N.; Chakravorty, D.; Zhang, Y.; Botella, J.R. Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. J. Plant Physiol. 2015, 188, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhao, Y.; Bi, G.; Liang, X.; Zhou, J.M. Early signalling mechanisms underlying receptor kinase-mediated immunity in plants. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180310. [Google Scholar] [CrossRef] [Green Version]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; He, P.; Shan, L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 2013, 110, 12114–12119. [Google Scholar] [CrossRef] [Green Version]
- Boisson-Dernier, A.; Franck, C.M.; Lituiev, D.S.; Grossniklaus, U. Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc. Natl. Acad. Sci. USA 2015, 112, 12211–12216. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Kärkönen, A.; Kuchitsu, K. Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 2015, 112, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.M.; Hazak, O.; Cheung, A.Y.; Yalovsky, S. RAC/ROP GTPases and auxin signaling. Plant Cell 2011, 23, 1208–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Ding, P.; Lian, K.; Wang, J.; Ma, M.; Li, L.; Li, M.; Zhang, X.; Chen, S.; Zhang, Y.; et al. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 2016, 5, e13568. [Google Scholar] [CrossRef]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Zulawski, M.; Schulze, G.; Braginets, R.; Hartmann, S.; Schulze, W.X. The Arabidopsis Kinome: Phylogeny and evolutionary insights into functional diversification. BMC Genom. 2014, 15, 548. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Kim, Y.J.; Kim, M.H.; Kwak, J.M. MAPK cascades in guard cell signal transduction. Front. Plant Sci. 2016, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Yamada, K.; Yoshimura, S.; Yamaguchi, K. Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant Signal. Behav. 2017, 12, e1361076. [Google Scholar] [CrossRef]
- Canales, C.; Bhatt, A.M.; Scott, R.; Dickinson, H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 2002, 12, 1718–1727. [Google Scholar] [CrossRef]
- Zhao, D.Z.; Wang, G.F.; Speal, B.; Ma, H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002, 16, 2021–2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Dickinson, H.G. Packaging the male germline in plants. Trends Genet. 2007, 23, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Liu, X.; Owen, H.A.; Zhao, D. Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 2220–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Li, Z.; Biener, G.; Xiong, E.; Malik, S.; Eaton, N.; Zhao, C.Z.; Raicu, V.; Kong, H.; Zhao, D. Carbonic anhydrases function in anther cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 2017, 29, 1335–1356. [Google Scholar] [CrossRef] [Green Version]
- Colcombet, J.; Boisson-Dernier, A.; Ros-Palau, R.; Vera, C.E.; Schroeder, J.I. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 2005, 17, 3350–3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Wang, Y.; Huang, J.; Ahsan, N.; Biener, G.; Paprocki, J.; Thelen, J.J.; Raicu, V.; Zhao, D. Two SERK receptor-like kinases interact with ems1 to control anther cell fate determination. Plant Physiol. 2017, 173, 326–337. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007, 50, 751–766. [Google Scholar] [CrossRef]
- Hord, C.L.; Chen, C.; Deyoung, B.J.; Clark, S.E.; Ma, H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 2006, 18, 1667–1680. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Hu, C.; Zhu, Y.; Cheng, K.; Li, X.; Wei, Z.; Xue, L.; Lin, F.; Shi, H.; Yi, J.; et al. CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 2018, 30, 2383–2401. [Google Scholar] [CrossRef] [Green Version]
- Hord, C.L.; Sun, Y.J.; Pillitteri, L.J.; Torii, K.U.; Wang, H.; Zhang, S.; Ma, H. Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 2008, 1, 645–658. [Google Scholar] [CrossRef]
- Wan, J.; Patel, A.; Mathieu, M.; Kim, S.Y.; Xu, D.; Stacey, G. A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 2008, 67, 469–482. [Google Scholar] [CrossRef]
- Huang, J.; Wijeratne, A.J.; Tang, C.; Zhang, T.; Fenelon, R.E.; Owen, H.A.; Zhao, D. Ectopic expression of TAPETUM DETERMINANT1 affects ovule development in Arabidopsis. J. Exp. Bot. 2016, 67, 1311–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, H.; Higashiyama, T. A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 2012, 10, e1001449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, H.; Higashiyama, T. Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 2016, 531, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, L.; Xue, Y.; Jia, P.F.; Chen, W.; Zhang, M.X.; Wang, Y.C.; Li, H.J.; Yang, W.C. Corrigendum: A receptor heteromer mediates the male perception of female attractants in plants. Nature 2016, 536, 360. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Restrepo, J.M.; Huck, N.; Kessler, S.; Gagliardini, V.; Gheyselinck, J.; Yang, W.C.; Grossniklaus, U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 2007, 317, 656–660. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Roy, S.; Kritsas, K.; Grobei, M.A.; Jaciubek, M.; Schroeder, J.I.; Grossniklaus, U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 2009, 136, 3279–3288. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.Y.; Shi, D.Q.; Jia, P.F.; Tang, J.; Li, H.J.; Liu, J.; Yang, W.C. The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Genet. 2016, 12, e1005933. [Google Scholar] [CrossRef] [Green Version]
- Nodine, M.D.; Yadegari, R.; Tax, F.E. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 2007, 12, 943–956. [Google Scholar] [CrossRef] [Green Version]
- Tsuwamoto, R.; Fukuoka, H.; Takahata, Y. GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 2008, 54, 30–42. [Google Scholar] [CrossRef]
- Tanaka, H.; Watanabe, M.; Sasabe, M.; Hiroe, T.; Tanaka, T.; Tsukaya, H.; Ikezaki, M.; Machida, C.; Machida, Y. Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 2007, 134, 1643–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, S.E. Organ formation at the vegetative shoot meristem. Plant Cell 1997, 9, 1067–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef]
- Lenhard, M.; Laux, T. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 2003, 130, 3163–3173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, H.; Zhu, Y.; Ma, Y.; Berkowitz, G.A. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca(2+) as a secondary cytosolic messenger. Plant J. 2016, 85, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Takemura, M.; Tani, E.; Nemoto, K.; Yokota, A.; Kohchi, T. An Arabidopsis MADS-box protein, AGL24, is specifically bound to and phosphorylated by meristematic receptor-like kinase (MRLK). Plant Cell Physiol. 2003, 44, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, R.; Takahashi, T.; Kato, A.; Torii, K.U.; Komeda, Y. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 1998, 15, 301–310. [Google Scholar] [CrossRef]
- Shpak, E.D.; Lakeman, M.B.; Torii, K.U. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 2003, 15, 1095–1110. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Kuroha, T.; Hnilova, M.; Khatayevich, D.; Kanaoka, M.M.; McAbee, J.M.; Sarikaya, M.; Tamerler, C.; Torii, K.U. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012, 26, 126–136. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Chen, X.; Mang, H.; Liu, C.; Yu, X.; Gao, X.; Torii, K.U.; He, P.; Shan, L. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 2015, 25, 2361–2372. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Liu, J.; Liu, Y.; Chen, L.; Sun, T.; Yao, N.; Wang, H.B.; Liu, B. BIK1 and ERECTA Play opposing roles in both leaf and inflorescence development in. Front. Plant Sci. 2019, 10, 1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, I.; Vassileva, V.; De Rybel, B.; Levesque, M.P.; Grunewald, W.; Van Damme, D.; Van Noorden, G.; Naudts, M.; Van Isterdael, G.; De Clercq, R.; et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 2008, 322, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, Y.; Grabowski, S.; Bleckmann, A.; Kuhnemuth, R.; Weidtkamp-Peters, S.; Pinto, K.G.; Kirschner, G.K.; Schmid, J.B.; Wink, R.H.; Hulsewede, A.; et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 2013, 23, 362–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelagio-Flores, R.; Muñoz-Parra, E.; Barrera-Ortiz, S.; Ortiz-Castro, R.; Saenz-Mata, J.; Ortega-Amaro, M.A.; Jiménez-Bremont, J.F.; López-Bucio, J. The cysteine-rich receptor-like protein kinase CRK28 modulates Arabidopsis growth and development and influences abscisic acid responses. Planta 2019, 251, 2. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.H.; Schiefelbein, J. A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 2008, 18, 1949–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, R.K.; Fulton, L.; Batoux, M.; Schneitz, K. The Arabidopsis receptor-like kinase STRUBBELIG mediates inter-cell-layer signaling during floral development. Dev. Biol. 2008, 323, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Vaddepalli, P.; Herrmann, A.; Fulton, L.; Oelschner, M.; Hillmer, S.; Stratil, T.F.; Fastner, A.; Hammes, U.Z.; Ott, T.; Robinson, D.G.; et al. The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 2014, 141, 4139–4148. [Google Scholar] [CrossRef] [Green Version]
- Kwak, S.H.; Schiefelbein, J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 2007, 302, 118–131. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Ishida, T.; Yamada, M.; Shigenobu, S.; Tabata, R.; Kinoshita, A.; Yamaguchi, K.; Hasebe, M.; Mitsumasu, K.; Sawa, S. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 2015, 208, 1104–1113. [Google Scholar] [CrossRef]
- Campos, R.; Goff, J.; Rodriguez-Furlan, C.; Van Norman, J.M. The Arabidopsis receptor kinase IRK is polarized and represses specific cell divisions in roots. Dev. Cell 2020, 52, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, K.; Turner, S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007, 17, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Kucukoglu, M.; Zhang, L.; Chen, P.; Decker, D.; Nilsson, O.; Jones, B.; Sandberg, G.; Zheng, B. The Arabidopsis LRR-RLK, PXC1, is a regulator of secondary wall formation correlated with the TDIF-PXY/TDR-WOX4 signaling pathway. BMC Plant Biol. 2013, 13, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gursanscky, N.R.; Jouannet, V.; Grunwald, K.; Sanchez, P.; Laaber-Schwarz, M.; Greb, T. MOL1 is required for cambium homeostasis in Arabidopsis. Plant J. 2016, 86, 210–220. [Google Scholar] [CrossRef] [Green Version]
- Bryan, A.C.; Obaidi, A.; Wierzba, M.; Tax, F.E. XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 2012, 235, 111–122. [Google Scholar] [CrossRef]
- Stenvik, G.E.; Butenko, M.A.; Aalen, R.B. Identification of a putative receptor-ligand pair controlling cell separation in plants. Plant Signal. Behav. 2008, 3, 1109–1110. [Google Scholar] [CrossRef] [Green Version]
- Taylor, I.; Wang, Y.; Seitz, K.; Baer, J.; Bennewitz, S.; Mooney, B.P.; Walker, J.C. Analysis of phosphorylation of the receptor-like protein kinase HAESA during Arabidopsis floral abscission. PLoS ONE 2016, 11, e0147203. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.; Brandt, B.; Wildhagen, M.; Hohmann, U.; Hothorn, L.A.; Butenko, M.A.; Hothorn, M. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 2016, 5, e15075. [Google Scholar] [CrossRef]
- Li, X.; Ahmad, S.; Ali, A.; Guo, C.; Li, H.; Yu, J.; Zhang, Y.; Gao, X.; Guo, Y. Characterization of somatic embryogenesis receptor-like kinase 4 as a negative regulator of leaf senescence in Arabidopsis. Cells 2019, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Wang, H.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, Y.; Kim, J.W.; Lee, H.S.; Lee, W.S.; Kim, S.K.; Wang, Z.Y.; Kim, S.H. Identification of Arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes. Plant Cell Physiol. 2013, 54, 1620–1634. [Google Scholar] [CrossRef] [Green Version]
- Wierzba, M.P.; Tax, F.E. An Allelic Series of bak1 Mutations Differentially Alter bir1 Cell Death, Immune Response, Growth, and Root Development Phenotypes in Arabidopsis thaliana. Genetics 2016, 202, 689–702. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Kumar, R.; Baek, D.; Hyun, T.K.; Chung, W.S.; Yun, D.J.; Kim, J.Y. Arabidopsis thaliana RECEPTOR DEAD KINASE1 functions as a positive regulator in plant responses to ABA. Mol. Plant 2017, 10, 223–243. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Zhang, G.; Zhou, Y.; Zhang, Z.; Wang, W.; Du, Y.; Wu, Z.; Song, C.P. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef]
- Corwin, J.A.; Kliebenstein, D.J. Quantitative resistance: More Than just perception of a pathogen. Plant Cell 2017, 29, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Chinchilla, D.; Boller, T.; Robatzek, S. Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 2007, 598, 358–371. [Google Scholar] [CrossRef]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tor, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef] [Green Version]
- Segonzac, C.; Zipfel, C. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 2011, 14, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Halter, T.; Imkampe, J.; Blaum, B.S.; Stehle, T.; Kemmerling, B. BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal. Behav. 2014, 9, e28944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaum, B.S.; Mazzotta, S.; Noldeke, E.R.; Halter, T.; Madlung, J.; Kemmerling, B.; Stehle, T. Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 2014, 186, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Kemmerling, B.; Schwedt, A.; Rodriguez, P.; Mazzotta, S.; Frank, M.; Qamar, S.A.; Mengiste, T.; Betsuyaku, S.; Parker, J.E.; Mussig, C.; et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 2007, 17, 1116–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Schwessinger, B.; Roux, M.; Kadota, Y.; Ntoukakis, V.; Sklenar, J.; Jones, A.; Zipfel, C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011, 7, e1002046. [Google Scholar] [CrossRef] [Green Version]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Pearce, G.; Ryan, C.A. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 2006, 103, 10104–10109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postel, S.; Kufner, I.; Beuter, C.; Mazzotta, S.; Schwedt, A.; Borlotti, A.; Halter, T.; Kemmerling, B.; Nurnberger, T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 2010, 89, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, G.; Shi, H.; Tang, D. RECEPTOR-LIKE KINASE 902 associates with and phosphorylates BRASSINOSTEROID-SIGNALING KINASE1 to regulate plant immunity. Mol. Plant 2019, 12, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Ntoukakis, V.; Rathjen, J.P. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 2009, 4, 539–541. [Google Scholar] [CrossRef]
- Mélida, H.; Sopeña-Torres, S.; Bacete, L.; Garrido-Arandia, M.; Jordá, L.; López, G.; Muñoz-Barrios, A.; Pacios, L.F.; Molina, A. Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 2018, 93, 34–49. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.X.; Li, C.L.; Xie, Z.P.; Staehelin, C. LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 5507–5516. [Google Scholar] [CrossRef]
- Chiu, C.H.; Paszkowski, U. Receptor-like kinases sustain symbiotic scrutiny. Plant Physiol. 2020, 182, 1597–1612. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.R.; Pandey, S. Specific subunits of heterotrimeric G proteins play important roles during nodulation in soybean. Plant Physiol. 2013, 162, 522–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Ding, P.; Sun, T.; Nitta, Y.; Dong, O.; Huang, X.; Yang, W.; Li, X.; Botella, J.R.; Zhang, Y. Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 2013, 161, 2146–2158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohorn, B.D.; Hoon, D.; Minkoff, B.B.; Sussman, M.R.; Kohorn, S.L. Rapid oligo-galacturonide induced changes in protein phosphorylation in Arabidopsis. Mol. Cell. Proteom. 2016, 15, 1351–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franck, C.M.; Westermann, J.; Boisson-Dernier, A. Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 2018, 69, 301–328. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca. Curr. Biol. 2018, 28, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G.; et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef] [Green Version]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Van der Does, D.; Zipfel, C.; Hamann, T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018, 11, eaao3070. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Shan, L.; Lin, N.C.; Martin, G.B.; Kemmerling, B.; Nurnberger, T.; Sheen, J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 2006, 125, 563–575. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; He, P.; Li, J.; Heese, A.; Peck, S.C.; Nürnberger, T.; Martin, G.B.; Sheen, J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Wang, X.; Wang, D.; Xu, F.; Ding, X.; Zhang, Z.; Bi, D.; Cheng, Y.T.; Chen, S.; Li, X.; et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar] [CrossRef] [Green Version]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef] [PubMed]
- Van der Burgh, A.M.; Postma, J.; Robatzek, S.; Joosten, M.H.A.J. Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 2019, 20, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtake, Y.; Takahashi, T.; Komeda, Y. Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, Z. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000, 24, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 2001, 126, 473–476. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Han, X.; Shi, R.; Yang, G.; Qi, L.; Wang, R.; Li, G. Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 2013, 73, 383–391. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef]
- Chen, K.; Fan, B.; Du, L.; Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 2004, 56, 271–283. [Google Scholar] [CrossRef]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.M.; Park, O.K. The Arabidopsis cysteine-rich receptor-like kinase CRK36 Regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Sensing the environment: Key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 2013, 64, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Manabe, Y.; Bressan, R.A.; Wang, T.; Li, F.; Koiwa, H.; Sokolchik, I.; Li, X.; Maggio, A. The Arabidopsis kinase-associated protein phosphatase regulates adaptation to Na+ stress. Plant Physiol. 2008, 146, 612–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.L.; Liu, X.; et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E5519–E5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Mizuno, S.; Tanaka, H.; Maruyama, K.; Osakabe, K.; Todaka, D.; Fujita, Y.; Kobayashi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 2010, 285, 9190–9201. [Google Scholar] [CrossRef] [Green Version]
- Hua, D.; Wang, C.; He, J.; Liao, H.; Duan, Y.; Zhu, Z.; Guo, Y.; Chen, Z.; Gong, Z. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 2012, 24, 2546–2561. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Liang, S.; Wu, Z.; Bi, C.; Yu, Y.T.; Wang, X.F.; Zhang, D.P. Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 2016, 67, 5009–5027. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, C.G.; Hwang, S.G.; Park, Y.C.; Park, H.M.; Kim, D.S.; Park, D.H.; Jang, C.S. Molecular characterization of the cold- and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations. J. Plant Physiol. 2015, 176, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chaudhuri, S.; Yang, L.; Du, L.; Poovaiah, B.W. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 2010, 285, 7119–7126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Shad Ali, G.; Yang, L.; Du, L.; Reddy, A.S.; Poovaiah, B.W. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef] [PubMed]







| S. No. | RLK Type | Gene (s) | Function | Ref |
|---|---|---|---|---|
| 1 | LRR | CLAVATA1 | Meristem and organ development | [17] |
| SERK | Microsporogenesis, embryogenesis, and embryonic competence in tissue culture | [18] | ||
| HAESA | Floral organ abscission | [19] | ||
| FLS2 | Senses bacterial flagellin | [20] | ||
| 2 | LecRLK | LecRK1 | Oligosaccharide-mediated signal transduction | [33] |
| 3 | WAK-RLK | WAK1 | Cell wall integrity, pathogen response | [21] |
| 4 | Extensin | LRX1 | Root hair morphogenesis | [22] |
| 5 | PERK | PERK4 | Cell wall integrity and drought response | [23] |
| 6 | CrRLK | HERK1 | Determinants of pollen tube | [35] |
| FER | Polar growth of root hair and pollen tube | [24,25] | ||
| 7 | S-domain | AtS1 | Self-incompatibility | [34] |
| ARK2, ARK3 | Organ maturation | [34] | ||
| 8 | CR-like | ACR4 | Epidermal patterning, integument development in ovules | [26] |
| Plant defense | [27] | |||
| 9 | DUF26 | CRK13 | Biotic stress response | [28] |
| CRK6, CRK7 | Oxidative stress response | [29] | ||
| 10 | LysM-RLK | AtCERK1 | Perception of MAMPs | [30] |
| 11 | Thaumatin | PR5K | Response to pathogenic signals | [31] |
| 12 | LRK 10-like | LRK10L1.2 | Drought resistance | [32] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jose, J.; Ghantasala, S.; Roy Choudhury, S. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. Int. J. Mol. Sci. 2020, 21, 4000. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114000
Jose J, Ghantasala S, Roy Choudhury S. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. International Journal of Molecular Sciences. 2020; 21(11):4000. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114000
Chicago/Turabian StyleJose, Jismon, Swathi Ghantasala, and Swarup Roy Choudhury. 2020. "Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery" International Journal of Molecular Sciences 21, no. 11: 4000. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114000