Choline Supplementation Sensitizes Legionella dumoffii to Galleria mellonella Apolipophorin III
Abstract
:1. Introduction
2. Results
2.1. Effects of Apolipophorin III on the L. dumoffii Cell Surface
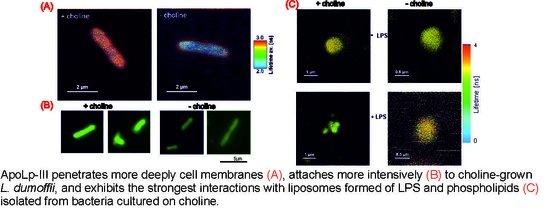
2.2. Interaction of Fluorescently Labeled Apolipophorin III with L. dumoffii Cells
2.3. Fluorescence Lifetime Imaging Microscopy (FLIM) Analysis of ApoLp-III Interaction with L. dumoffii Cells
2.4. Characteristics of L. dumoffii Lipopolysaccharide (LPS)
2.4.1. Electrophoretic Analysis of Lipopolysaccharide (LPS)
2.4.2. Fatty Acid Profile and Sugar Analyses
2.5. Interaction of Apolipophorin III with L. dumoffii Lipopolysaccharide (LPS)
2.6. FLIM Analysis of Apolipophorin III Interaction with Liposomes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Culture Conditions
4.2. Preparation of L. dumoffii Phospholipids
4.3. Isolation, Purification, and Electrophoretic Analysis of L. dumoffii Lipopolysaccharide (LPS)
4.4. Compositional Analyses of L. dumoffii Lipopolysaccharide (LPS)
4.4.1. Fatty Acid Analysis
4.4.2. Sugar Analysis
4.4.3. Gas Liquid Chromatography and Mass Spectrometry (GLC-MS)
4.5. Apolipophorin III Purification and Fluorescent Labeling
4.6. Interaction of Apolipophorin III with L. dumoffii Lipopolysaccharide (LPS)
4.7. Liposome Preparation, Apolipophorin III Interaction with Liposomes, and FLIM Measurements of Liposomes
4.7.1. Chemicals
4.7.2. Giant Unilamellar Vesicles
4.7.3. Small Unilamellar Vesicles
4.7.4. Microscopy Experiments
4.8. Scanning Electron Microscopy Imaging and X-ray Analysis of Bacterial Cells
4.9. Apolipophorin III Binding to L. dumoffii Cells and Imaging of Bacteria
4.9.1. Laser Scanning Confocal Microscopy
4.9.2. Fluorescence Lifetime Imaging Microscopy
4.9.3. Atomic Force Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | atomic force microscopy |
| apoLp-III | apolipophorin III |
| CAP | community-acquired pneumonia |
| FITC | fluorescein isothiocyanate |
| FLIM | fluorescence lifetime imaging microscopy |
| HMDS | hexamethyldisilane |
| HPLC | high-pressure liquid chromatography |
| LPS | lipopolysaccharide |
| LTA | lipoteichoic acid |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| SEC | size exclusion chromatography |
| SEM | scanning electron microscopy |
| TCSPC | time-correlated single photon counting |
| TFA | trifluoroacetic acid |
| TMCS | trimethylchlorosilane |
| TNF-α | tumour necrosis factor α |
References
- Craun, G.F.; Brunkard, J.M.; Yoder, J.S.; Roberts, V.A.; Carpenter, J.; Wade, T.; Calderon, R.L.; Roberts, J.M.; Beach, M.J.; Roy, S.L. Causes of outbreaks associated with drinking water in the United States from 1971 to 2006. Clin. Microbiol. Rev. 2010, 23, 507–528. [Google Scholar] [CrossRef] [Green Version]
- Dufresne, S.F.; Locas, M.C.; Duchesne, A.; Restieri, C.; Ismail, J.; Lefebvre, B.; Labbe, A.C.; Dion, R.; Plante, M.; Laverdiere, M. Sporadic Legionnaires’ disease: The role of domestic electric hot-water tanks. Epidemiol. Infect. 2012, 140, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Beauté, J. The European Legionnaires’ Disease Surveillance Network. Legionnaires’ disease in Europe, 2011 to 2015. Euro Surveill. 2017, 22, 30566. [Google Scholar] [CrossRef] [Green Version]
- Garrison, L.E.; Kunz, J.M.; Cooley, L.A.; Moore, M.R.; Lucas, C.; Schrag, S.; Sarisky, J.; Whitney, C.G. Vital signs: Deficiencies in environmental control identified in outbreaks of Legionnaires’ disease—North America, 2000–2014. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 576–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez, A.; Alvarez, J.; Sabria, M.; Carmona, G.; Torner, N.; Oviedo, M.; Cayla, J.; Minguell, S.; Barrabeig, I.; Sala, M.; et al. Factors influencing the case-fatality rate of Legionnaires’ disease. Int. J. Tuberc. Lung. Dis. 2009, 13, 407–412. [Google Scholar]
- Yu, L.V.; Plouffe, F.J.; Pastoris, M.C.; Stout, J.E.; Schousboe, M.; Widmer, A.; Summersgill, J.; File, T.; Heath, C.M.; Paterson, D.L.; et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J. Infect. Dis. 2002, 186, 127–128. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control. European Legionnaires’ Disease Surveillance Network (ELDSNet)—Operating Procedures for the Surveillance of Travel-Associated Legionnaires’ Disease in the EU/EEA; ECDC: Solna Municipality, Sweden, 2017. [Google Scholar] [CrossRef]
- Bartram, J.; Chartier, Y.; Lee, J.V.; Pond, K.; Surman-Lee, S. Legionella and the prevention of legionellosis. World Health Organ. 2007. [Google Scholar] [CrossRef]
- Maruta, K.; Miyamoto, H.; Hamada, T.; Ogawa, M.; Taniguchi, H.; Yoshida, S. Entry and intracellular growth of Legionella dumoffii in alveolar epithelial cells. Am. J. Respir. Crit. Care Med. 1998, 157, 1967–1974. [Google Scholar] [CrossRef] [PubMed]
- Fujita, I.H.; Tsuboi, M.; Ohotsuka, I.; Sano, Y.; Murakami, H.; Akioka, H. Legionella dumoffii and Legionella pneumophila serogroup 5 isolated from 2 cases of fulminant pneumonia. J. Jpn. Assoc. Infect. Dis. 1989, 63, 778–780. [Google Scholar] [CrossRef] [Green Version]
- Tompkins, L.S.; Roessler, B.J.; Redd, S.C.; Markowitz, L.E.; Cohen, M.L. Legionella prosthetic-valve endocarditis. N. Engl. J. Med. 1988, 318, 530–535. [Google Scholar] [CrossRef]
- Flendrie, M.; Jeurissen, M.; Franssen, M.; Kwa, D.; Klaassen, C.; Vos, F. Septic arthritis caused by Legionella dumoffii in a patient with systemic lupus erythematosus-like disease. J. Clin. Microbiol. 2011, 49, 746–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caroff, M.; Novikov, A. LPS structure, function, and heterogeneity. In Endotoxin Detection and Control in Pharma, Limulus, and Mammalian Systems; Williams, K.L., Ed.; Springer Nature: Berlin, Germany, 2019. [Google Scholar] [CrossRef]
- Geiger, O.; Lopez-Lara, I.M.; Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta 2013, 1831, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Conover, G.M.; Martinez-Morales, F.; Heidtman, M.I.; Luo, Z.Q.; Tang, M.; Chen, C.; Geiger, O.; Isberg, R.R. Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell. Microbiol. 2008, 10, 514–528. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Morales, F.; Schobert, M.; Lopez-Lara, I.M.; Geiger, O. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology 2003, 149, 3461–3471. [Google Scholar] [CrossRef]
- Palusinska-Szysz, M.; Janczarek, M.; Kalitynski, R.; Dawidowicz, A.L.; Russa, R. Legionella bozemanae synthesizes phosphatidylcholine from exogenous choline. Microbiol. Res. 2011, 166, 87–98. [Google Scholar] [CrossRef]
- Palusinska-Szysz, M.; Szuster-Ciesielska, A.; Kania, M.; Janczarek, M.; Chmiel, E.; Danikiewicz, W. Legionella dumoffii utilizes exogenous choline for phosphatidylcholine synthesis. Int. J. Mol. Sci. 2014, 15, 8256–8279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palusińska-Szysz, M.; Szuster-Ciesielska, A.; Janczarek, M.; Wdowiak-Wróbel, S.; Schiller, J.; Reszczyńska, E.; Gruszecki, W.I.; Fuchs, B. Genetic diversity of Legionella pcs and pmtA genes and the effect of utilization of choline by Legionella spp. on induction of proinflammatory cytokines. Pathog. Dis. 2019, 77. [Google Scholar] [CrossRef]
- Weers, P.M.M.; Ryan, R.O. Apolipophorin III: Role model apolipoprotein. Insect Biochem. Mol. Biol. 2006, 36, 231–240. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Cytryńska, M. Apolipophorins in insects immune response. Invertebr. Surv. J. 2013, 10, 58–68. [Google Scholar]
- Leon, L.J.; Pratt, C.C.; Vasquez, L.J.; Weers, P.M.M. Tyrosine fluorescence analysis of apolipophorin III—Lipopolysaccharide interaction. Archiv. Biochem. Biophys. 2006, 452, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Halwani, A.E.; Niven, D.F.; Dunphy, G.B. Apolipophorin-III and the interactions of lipoteichoic acids with the immediate immune responses of Galleria mellonella. J. Invertebr. Pathol. 2000, 76, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Pratt, C.C.; Weers, P.M.M. Lipopolysaccharide binding of an exchangeable apolipoprotein, apolipophorin III, from Galleria mellonella. Biol. Chem. 2004, 385, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Oztug, M.; Martinon, D.; Weers, P.M.M. Characterization of the apoLp-III/LPS complex: Insight into the mode of binding interaction. Biochemistry 2012, 51, 6220–6227. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.M.A.; Tew, I.F.; Lee, B.L.; Ratcliffe, N.A. A novel role for an insect apolipoprotein (apolipophorin III) in β-1,3-glucan pattern recognition and cellular encapsulation reactions. J. Immunol. 2004, 172, 2177–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niere, M.; Dettloff, M.; Maier, T.; Ziegler, M.; Wiesner, A. Insect immune activation by apolipophorin III is correlated with the lipid-binding properties of this protein. Biochemistry 2001, 40, 11502–11508. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Sowa-Jasiłek, A.; Stączek, S.; Jakubowicz, T.; Cytryńska, M. Different forms of apolipophorin III in Galleria mellonella larvae challenged with bacteria and fungi. Peptides 2015, 68, 105–112. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Cytryńska, M. Involvement of apolipophorin III in antibacterial defense of Galleria mellonella larvae. Comp. Biochem. Physiol. B 2011, 158, 90–98. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Stączek, S.; Mak, P.; Piersiak, T.; Skrzypiec, K.; Cytryńska, M. The effect of Galleria mellonella apolipophorin III on yeasts and filamentous fungi. J. Insect Physiol. 2012, 58, 164–177. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Januszanis, B.; Mak, P.; Cytryńska, M. An atomic force microscopy study of Galleria mellonella apolipophorin III effect on bacteria. Biochim. Biophys. Acta 2011, 1808, 1896–1906. [Google Scholar] [CrossRef] [Green Version]
- Palusinska-Szysz, M.; Zdybicka-Barabas, A.; Pawlikowska-Pawlęga, B.; Mak, P.; Cytryńska, M. Anti-Legionella dumoffii activity of Galleria mellonella defensin and apolipophorin III. Int. J. Mol. Sci. 2012, 13, 17048–17064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chmiel, E.; Palusinska-Szysz, M.; Zdybicka-Barabas, A.; Cytryńska, M.; Mak, P. The effect of Galleria mellonella hemolymph polypeptides on Legionella gormanii. Acta Biochim. Pol. 2014, 61, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Palusinska-Szysz, M.; Gruszecki, W.I.; Mak, P.; Cytryńska, M. Galleria mellonella apolipophorin III—An apolipoprotein with anti-Legionella pneumophila activity. Biochim. Biophys. Acta 2014, 1838, 2689–2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palusinska-Szysz, M.; Zdybicka-Barabas, A.; Cytryńska, M.; Wdowiak-Wróbel, S.; Chmiel, E.; Gruszecki, W.I. Analysis of cell surface alterations in Legionella pneumophila cells treated with human apolipoprotein E. Pathog. Dis. 2015, 73, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Palusińska-Szysz, M.; Zdybicka-Barabas, A.; Reszczyńska, E.; Luchowski, R.; Kania, M.; Gisch, N.; Waldow, F.; Mak, P.; Danikiewicz, W.; Gruszecki, W.I.; et al. The lipid composition of Legionella dumoffii membrane modulates the interaction with Galleria mellonella apolipophorin III. Biochim. Biophys. Acta 2016, 1861, 617–629. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Epand, R.F.; Rosenfeld, Y.; Peleg, A.; Barra, D.; Epand, R.M.; Shai, Y. Lipopolysaccharide, a key molecule involved in the synergism between temporins in inhibiting bacterial growth and in endotoxin neutralization. J. Biol. Chem. 2008, 283, 22907–22917. [Google Scholar] [CrossRef] [Green Version]
- Grudzinski, W.; Nierzwicki, R.; Welc, R.; Reszczynska, E.; Luchowski, R.; Czub, J.; Gruszecki, W.I. Localization and orientation of xanthophylls in a lipid bilayer. Sci. Rep. 2017, 7, 9619. [Google Scholar] [CrossRef]
- Zhang, Y.; Lewis, R.N.; McElhaney, R.N.; Ryan, R.O. Calorimetric and spectroscopic studies of the interaction of Manduca Sexta apolipophorin III with zwitterionic, anionic, and nonionic lipids. Biochemistry 1993, 32, 3942–3952. [Google Scholar] [CrossRef]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [Green Version]
- Verdon, J.; Labanowski, J.; Sahar, T.; Ferreira, T.; Lacombe, C.; Buchrieser, C.; Berjeaud, J.M.; Hechard, Y. Fatty acid composition modulates sensitivity of Legionella pneumophila to warnericin RK, an antimicrobial peptide. Biochim. Biophys. Acta 2011, 1808, 1146–1153. [Google Scholar] [CrossRef]
- Shevchuk, O.; Jäger, J.; Steinert, M. Virulence properties of the Legionella pneumophila cell envelope. Front. Microbiol. 2011, 2, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jäger, J.; Keese, S.; Roessle, M.; Steinert, M.; Schromm, A.B. Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell. Microbiol. 2015, 17, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Jäger, J.; Marwitz, S.; Tiefenau, J.; Rasch, J.; Shevchuk, O.; Kugler, C.; Goldmann, T.; Steinert, M. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect. Immun. 2014, 82, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urashima, A.; Sanou, A.; Yen, H.; Tobe, T. Enterohaemorrhagic Escherichia coli produces outer membrane vesicles as an active defence system against antimicrobial peptide LL-37. Cell. Microbiol. 2017, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Amemura-Maekawa, J.; Hayakawa, Y.; Sugie, H.; Moribayashi, A.; Kura, F.; Chang, B.; Wada, A.; Watanabe, H. Legioliulin, a new isocoumarin compound responsible for blue-white autofluorescence in Legionella (Fluoribacter) dumoffii under long-wavelength UV light. Biochem. Biophys. Res. Commun. 2004, 323, 954–959. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharide. Extraction with phenol-water and further applications of the procedure. Meth. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G.G. Tricine-sodium dodecyl sulphate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSxM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]







| Bacteria Culture Conditions (−/+Choline) and Treatment (−/+ ApoLp-III) | ||||
|---|---|---|---|---|
| −Choline | +Choline | |||
| −apoLp-III | +apoLp-III | −apoLp-III | +apoLp-III | |
| DMT modulus (MPa) | 1186 (±396.46) a | 7414.5 (±2334.21) A,* | 2538 (±790.12) b | 2067 (±569.12) B |
| Adhesion (nN) | 0.949 (±0.03) | 1.18 (±0.28) * | 0.958 (±0.05) | 1.146 (±0.10) * |
| RMS Roughness (nm) | 2.293 (±0.77) | 3.076 (±1.04) ** | 2.058 (±0.63) | 3.172 (±1.27) ** |
| Elements | Surface of L. dumoffii | OMV | ||
|---|---|---|---|---|
| Weight % | Atom % | Weight % | Atom % | |
| C | 52.49 ± 35.85 | 58.06 ± 34.89 | 27.77 ± 6.88 | 33.30 ± 7.37 |
| N | 2.90 ± 14 | 2.96 ± 0.05 | 7.66 ± 2.34 | 7.95 ± 2.34 |
| O | 43.23 ± 34.28 | 8.42 ± 32.92 | 63.13 ± 8.11 | 58.01 ± 9.13 |
| P | 0.66 ± 0.12 | 0.29 ± 0.25 | 1.18 ± 0.87 | 0.56 ± 0.43 |
| Cl | 0.72 ± 0.28 | 0.27 ± 0.80 | 0.26 ± 0.6 | 0.18 ± 0.42 |
| No | Retention Time | Fatty Acid | Relative Content (%) |
|---|---|---|---|
| 1 | 11.23 | 14:0 | 1 |
| 2 | 11.55 | 3-OH 12:0 | tr |
| 3 | 12.76 | 15:0 | 2 |
| 4 | 13.63 | 3-OH 13:0 | tr |
| 5 | 14.69 | 16:0 | 6 |
| 6 | 14.88 | i3-OH 14:0 | 1 |
| 7 | 15.44 | 16:0 | 9 |
| 8 | 15.62 | n3-OH 14:0 | 10 |
| 9 | 16.87 | 17:0 | 8 |
| 10 | 16.99 | a3-OH 15:0 | 3 |
| 11 | 17.53 | n3-OH 15:0 | 2 |
| 12 | 18.69 | i3-OH 16:0 | 2 |
| 13 | 19.35 | n3-OH 16:0 | 8 |
| 14 | 20.46 | i3-OH 17:0 | tr |
| 15 | 20.62 | a3-OH 17:0 | tr |
| 16 | 21.11 | n3-OH 17:0 | 1 |
| 17 | 22.16 | i3-OH 18:0 | tr |
| 18 | 22.77 | a3-OH 18:0 | 9 |
| 19 | 23.78 | n3-OH 19:0 | 0.5 |
| 20 | 23.95 | a3-OH 19:0 | 1 |
| 21 | 24.38 | n3-OH 19:0 | 2 |
| 22 | 25.92 | 3-OH 20:0 | 5 |
| 23 | 27.40 | 3-OH 21:0 | tr |
| 24 | 37.06 | 28:0 (27-oxo) | 17 |
| 25 | 37.17 | 27:0-dioic | 10 |
| 26 | 39.93 | 30:0-dioic | 3 |
| Retention Time | Component | Relative Content (%) |
|---|---|---|
| 18.18 | Quinovosamine | 56 |
| 19.27 | Mannose | 6 |
| 19.43 | Glucose | 2 |
| 19.61 | Galactose | 8 |
| 22.27 | Glucosamine | 17 |
| 22.64 | Galactosamine | 10 |
| 23.87 | 2,3-diamino 2,3-dideoxy-d-glucose | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palusińska-Szysz, M.; Zdybicka-Barabas, A.; Luchowski, R.; Reszczyńska, E.; Śmiałek, J.; Mak, P.; Gruszecki, W.I.; Cytryńska, M. Choline Supplementation Sensitizes Legionella dumoffii to Galleria mellonella Apolipophorin III. Int. J. Mol. Sci. 2020, 21, 5818. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165818
Palusińska-Szysz M, Zdybicka-Barabas A, Luchowski R, Reszczyńska E, Śmiałek J, Mak P, Gruszecki WI, Cytryńska M. Choline Supplementation Sensitizes Legionella dumoffii to Galleria mellonella Apolipophorin III. International Journal of Molecular Sciences. 2020; 21(16):5818. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165818
Chicago/Turabian StylePalusińska-Szysz, Marta, Agnieszka Zdybicka-Barabas, Rafał Luchowski, Emilia Reszczyńska, Justyna Śmiałek, Paweł Mak, Wiesław I. Gruszecki, and Małgorzata Cytryńska. 2020. "Choline Supplementation Sensitizes Legionella dumoffii to Galleria mellonella Apolipophorin III" International Journal of Molecular Sciences 21, no. 16: 5818. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165818