A Dynamic, Split-Luciferase-Based Mini-G Protein Sensor to Functionally Characterize Ligands at All Four Histamine Receptor Subtypes
Abstract
:1. Introduction
2. Results
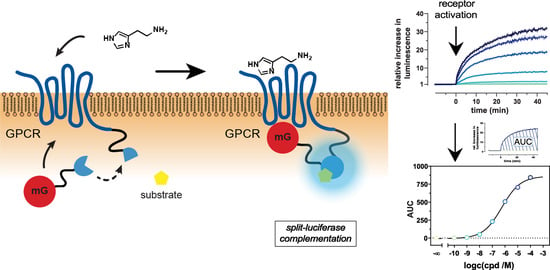
2.1. Principle and Characteristics of the Mini-G Protein Recruitment Assay
2.2. Kinetics and Dynamic Ranges of Mini G Protein Recruitment
2.3. Mini-G Protein Recruitment-Based Investigation of Histamine Receptor Ligands with Diverse Pharmacological Profiles
| Compound | Mini G Protein Recruitment | GTPγS/GTPase ‡ | Competition Binding | ||
|---|---|---|---|---|---|
| pEC50/(pKb) | Emax [%] | pEC50/(pKb) | Emax [%] | pKi | |
| his | 6.16 ± 0.09 | 100 | 5.21 ± 0.06 a 6.92 ‡,b | 100 a 100 ‡,b | 5.62 h |
| KUM530 | 6.41 ± 0.12 | 112 ± 1.0 * | 6.22 ± 0.10 a 7.75 ‡,c | 95 ± 5.7 94 ‡c | 6.43 j |
| betahis | 5.49 ± 0.13 | 75 ± 2.0 | 5.84 ‡,d | 86 ‡,d | |
| histapro | 6.39 ± 0.03 | 33 ± 2.0 | 5.86 ± 0.07 a 6.95 ‡,b | 31 ± 2.8 62 ‡,b | 6.47 h |
| Namh | 5.56 ± 0.08 | 99 ± 2.0 | |||
| 4mhis | 4.46 ± 0.16 | 44 ± 2.4 | 4.80 ‡,e | 90 ‡,e | |
| PI294 | 4.93 ± 0.03 | 29 ± 1.4 | 5.46 ‡,f | 30 ‡,f | |
| suprahis | 6.09 ± 0.13 | 49 ± 3.7 | 6.83 ‡,b | 64 ‡,b | 6.58 h |
| dph | 6.95 ± 0.04 (6.69) ± 0.17 | −4 ± 0.1 | (6.98) ± 0.07 a (7.81) ‡,d | 7.40 k | |
| map | 8.51 ± 0.04 (10.58) ± 0.11 | −4 ± 0.2 | (8.54) ‡,g | 8.50 k | |
| mep | 8.36 ± 0.11 (8.54) ± 0.19 | −3 ± 0.2 | (8.00) ± 0.17 a (8.25) ‡,d | 8.39 k 8.7 l | |
| cyp | 8.68 ± 0.24 (10.19) ± 0.10 | −3 ± 0.5 | (8.72) ‡,d | 8.63 k | |
| Compound | Mini G Protein Recruitment | GTPase | Competition Binding | ||
|---|---|---|---|---|---|
| pEC50/(pKb) | Emax [%] | pEC50/(pKb) | Emax [%] | pKi | |
| his | 6.94 ± 0.05 | 100 | 6.00 a | 100 a | 6.27 d |
| impro | 7.48 ± 0.01 | 90 ± 1.5 | 6.80 a | 82 a | 6.3 e |
| amt | 7.57 ± 0.08 | 105 ± 2.8 | 6.72 a | 85 a | 6.61 d |
| dim | 6.47 ± 0.04 | 94 ± 2.6 | 6.04 a | 91 a | 4.6 e |
| Namh | 6.76 ± 0.09 | 93 ± 1.7 | |||
| 4mhis | 6.37 ± 0.05 | 93 ± 2.2 | 5.54 b | 101 b | 5.1 f |
| PI294 | 6.92 ± 0.13 | 95 ± 1.1 | 6.43 c | 83 c | |
| cim | 6.02 ± 0.04 (6.28) ± 0.02 | −8 ± 0.8 | (5.77) a | −8 a | 6.2 e |
| fam | 7.29 ± 0.10 (8.14) ± 0.09 | −9 ± 0.7 | (7.32) a | −1 a | 7.8 e 6.87 d |
| ran | 7.02 ± 0.11 (6.99) ± 0.01 | −8 ± 0.7 | (6.08) a | −9 a | 7.1 e 5.76 d |
| Compound | Mini G Protein Recruitment | GTPγS/GTPase ‡ | Competition Binding | ||
|---|---|---|---|---|---|
| pEC50/(pKb) | Emax [%] | pEC50/(pKb) | Emax [%] | pKi/(pKd) | |
| his | 6.47 ± 0.04 | 100 | 7.3 a | 89 a | 7.96 f |
| imet | 8.30 ± 0.17 | 67 ± 0.7 | 8.6 a | 80 a | 8.8 g |
| immep | 8.77 ± 0.05 | 63 ± 1.3 | 8.8 a | 77 a | 9.3 g |
| VUF8430 | 5.21 ± 0.12 | 43 ± 1.6 | 6.0 h | ||
| Namh | 7.20 ± 0.03 | 111 ± 1.6 * | 7.9 a | 100 a | 8.4 g |
| 4mhis | 4.53 ± 0.08 | 19 ± 1.5 | |||
| PI294 | 8.40 ± 0.06 | 11 ± 1.1 | 8.80 ‡,b | 39 ‡,b | (8.96) j |
| thio | 7.41 ± 0.04 (7.21) ± 0.07 | −3 ± 0.4 | 6.9 a | −52 a | 7.42 f |
| clob | 9.05 ± 0.10 (9.28) ± 0.12 | −3 ± 0.2 | 9.14 ‡,c (9.28) d | −137 ‡,c | 9.34 f |
| JNJ | (5.44) ± 0.01 | 5.29 k | |||
| pito | (8.41) ± 0.05 | (9.80) e | 8.57 l | ||
| Compound | Mini G Protein Recruitment | GTPγS/GTPase ‡ | Competition Binding | ||
|---|---|---|---|---|---|
| pEC50/(pKb) | Emax [%] | pEC50/(pKb) | Emax [%] | pKi/(pKd) | |
| his | 6.40 ± 0.04 | 100 | 7.60 ‡,a | 100 ‡,a | 7.8 f |
| imet | 6.94 ± 0.04 | 47 ± 0.1 | 8.17 ‡,b | 69 ‡,b | 8.2 f |
| immep | 6.73 ± 0.05 | 66 ± 2.8 | 7.35 ‡,b | 68 ‡,b | 7.7 f |
| VUF8430 | 6.47 ± 0.03 | 60 ± 0.2 | 7.42 c | 84 c | 7.5 f |
| Namh | 5.68 ± 0.06 | 82 ± 1.1 | 6.5 f | ||
| 4mhis | 6.48 ± 0.06 | 78 ± 0.5 | 7.15 ‡,d | 90 ‡,d | 7.30 f |
| PI294 | 7.71 ± 0.04 | 85 ± 0.6 | 8.35 c | 102 c | (8.29) g |
| clob | 7.28 ± 0.06 | 48 ± 2.0 | 7.65 c | 45 c | 7.75 h |
| thio | 6.68 ± 0.04 (6.90) ± 0.01 | −8 ± 1.9 | 6.58 c (6.83) c | −139 c | 6.9 e |
| JNJ | (7.25) ± 0.25 | 0 to 2.8 | 7.10 c (7.60) c | −39 c | 7.52 h |
| A943931 | (8.43) ± 0.22 | −4 to 2.8 | 7.3 e | −180 e | 8.33 j |
2.4. Influence of Mini-G Protein Co-Expression on Potencies and Dynamic Ranges
2.5. Stabilization of the Active H2R Conformation by the Minimal Gαs Protein
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Molecular Cloning
4.3. Cell Culture
4.4. Generation of Stable Transfectants
4.5. Generation of Transient Transfectants
4.6. Western Blot Analysis
4.7. Mini-G Protein Recruitment Assay
4.8. Radioligand Binding Experiments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| H1R | Human histamine H1 receptor |
| H2R | Human histamine H2 receptor |
| H3R | Human histamine H3 receptor |
| H4R | Human histamine H4 receptor |
| HEPES | 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid |
| L-15 | Leibovitz’ L-15 medium |
| NanoLuc | NanoLuc luciferase |
References
- Hamm, H.E. How activated receptors couple to g proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 4819–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, G.; Kostenis, E. Heterotrimeric g-proteins: A short history. Br. J. Pharmacol. 2006, 147 (Suppl. 1), S46–S55. [Google Scholar] [CrossRef] [Green Version]
- Gutowski, S.; Smrcka, A.; Nowak, L.; Wu, D.G.; Simon, M.; Sternweis, P.C. Antibodies to the alpha q subfamily of guanine nucleotide-binding regulatory protein alpha subunits attenuate activation of phosphatidylinositol 4,5-bisphosphate hydrolysis by hormones. J. Biol. Chem. 1991, 266, 20519–20524. [Google Scholar] [PubMed]
- Dowal, L.; Provitera, P.; Scarlata, S. Stable association between g alpha(q) and phospholipase c beta 1 in living cells. J. Biol. Chem. 2006, 281, 23999–24014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunahara, R.K.; Insel, P.A. The molecular pharmacology of g protein signaling then and now: A tribute to alfred g. Gilman. Mol. Pharmacol. 2016, 89, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazar, A.M.; Irannejad, R.; Baldwin, T.A.; Sundaram, A.B.; Gutkind, J.S.; Inoue, A.; Dessauer, C.W.; Von Zastrow, M. G protein-regulated endocytic trafficking of adenylyl cyclase type 9. Elife 2020, 9, e58039. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in gpcr drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Schnell, D.; Burleigh, K.; Trick, J.; Seifert, R. No evidence for functional selectivity of proxyfan at the human histamine h3 receptor coupled to defined gi/go protein heterotrimers. J. Pharmacol. Exp. Ther. 2010, 332, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- Wifling, D.; Loffel, K.; Nordemann, U.; Strasser, A.; Bernhardt, G.; Dove, S.; Seifert, R.; Buschauer, A. Molecular determinants for the high constitutive activity of the human histamine h4 receptor: Functional studies on orthologues and mutants. Br. J. Pharmacol. 2015, 172, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Bongers, G.; Krueger, K.M.; Miller, T.R.; Baranowski, J.L.; Estvander, B.R.; Witte, D.G.; Strakhova, M.I.; van Meer, P.; Bakker, R.A.; Cowart, M.D.; et al. An 80-amino acid deletion in the third intracellular loop of a naturally occurring human histamine h3 isoform confers pharmacological differences and constitutive activity. J. Pharmacol. Exp. Ther. 2007, 323, 888–898. [Google Scholar] [CrossRef] [Green Version]
- Preuss, H.; Ghorai, P.; Kraus, A.; Dove, S.; Buschauer, A.; Seifert, R. Constitutive activity and ligand selectivity of human, guinea pig, rat, and canine histamine h2 receptors. J. Pharmacol. Exp. Ther. 2007, 321, 983–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, A.; Striegl, B.; Wittmann, H.J.; Seifert, R. Pharmacological profile of histaprodifens at four recombinant histamine h1 receptor species isoforms. J. Pharmacol. Exp. Ther. 2008, 324, 60–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, R.; Wenzel-Seifert, K.; Gether, U.; Kobilka, B.K. Functional differences between full and partial agonists: Evidence for ligand-specific receptor conformations. J. Pharmacol. Exp. Ther. 2001, 297, 1218–1226. [Google Scholar] [PubMed]
- Koval, A.; Kopein, D.; Purvanov, V.; Katanaev, V.L. Europium-labeled gtp as a general nonradioactive substitute for [(35)s]gtpgammas in high-throughput g protein studies. Anal. Biochem. 2010, 397, 202–207. [Google Scholar] [CrossRef]
- Anderson, E.K.; Martin, D.S. A fluorescent gtp analog as a specific, high-precision label of microtubules. Biotechniques 2011, 51, 43–48. [Google Scholar] [CrossRef]
- Mondal, S.; Hsiao, K.; Goueli, S.A. A homogenous bioluminescent system for measuring gtpase, gtpase activating protein, and guanine nucleotide exchange factor activities. Assay Drug Dev. Technol. 2015, 13, 444–455. [Google Scholar] [CrossRef] [Green Version]
- Strange, P.G. Use of the gtpgammas ([35s]gtpgammas and eu-gtpgammas) binding assay for analysis of ligand potency and efficacy at g protein-coupled receptors. Br. J. Pharmacol. 2010, 161, 1238–1249. [Google Scholar] [CrossRef] [Green Version]
- Denis, C.; Sauliere, A.; Galandrin, S.; Senard, J.M.; Gales, C. Probing heterotrimeric g protein activation: Applications to biased ligands. Curr. Pharm. Des. 2012, 18, 128–144. [Google Scholar] [CrossRef] [Green Version]
- Hein, P.; Frank, M.; Hoffmann, C.; Lohse, M.J.; Bunemann, M. Dynamics of receptor/g protein coupling in living cells. EMBO J. 2005, 24, 4106–4114. [Google Scholar] [CrossRef] [Green Version]
- Gales, C.; Van Durm, J.J.; Schaak, S.; Pontier, S.; Percherancier, Y.; Audet, M.; Paris, H.; Bouvier, M. Probing the activation-promoted structural rearrangements in preassembled receptor-g protein complexes. Nat. Struct. Mol. Biol. 2006, 13, 778–786. [Google Scholar] [CrossRef]
- Mocking, T.A.M.; Buzink, M.; Leurs, R.; Vischer, H.F. Bioluminescence resonance energy transfer based g protein-activation assay to probe duration of antagonism at the histamine h3 receptor. Int. J. Mol. Sci. 2019, 20, 3724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehme, R.; Carpenter, B.; Singhal, A.; Strege, A.; Edwards, P.C.; White, C.F.; Du, H.; Grisshammer, R.; Tate, C.G. Mini-g proteins: Novel tools for studying gpcrs in their active conformation. PLoS ONE 2017, 12, e0175642. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Okashah, N.; Inoue, A.; Nehme, R.; Carpenter, B.; Tate, C.G.; Lambert, N.A. Mini g protein probes for active g protein-coupled receptors (gpcrs) in live cells. J. Biol. Chem. 2018, 293, 7466–7473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wouters, E.; Marin, A.R.; Dalton, J.A.R.; Giraldo, J.; Stove, C. Distinct dopamine d(2) receptor antagonists differentially impact d(2) receptor oligomerization. Int. J. Mol. Sci. 2019, 20, 1686. [Google Scholar] [CrossRef] [Green Version]
- Wouters, E.; Walraed, J.; Robertson, M.J.; Meyrath, M.; Szpakowska, M.; Chevigne, A.; Skiniotis, G.; Stove, C. Assessment of biased agonism among distinct synthetic cannabinoid receptor agonist scaffolds. ACS Pharmacol. Transl. Sci. 2020, 3, 285–295. [Google Scholar] [CrossRef]
- Pottie, E.; Tosh, D.K.; Gao, Z.G.; Jacobson, K.A.; Stove, C.P. Assessment of biased agonism at the a3 adenosine receptor using beta-arrestin and minigalphai recruitment assays. Biochem. Pharmacol. 2020, 177, 113934. [Google Scholar] [CrossRef]
- Pottie, E.; Dedecker, P.; Stove, C.P. Identification of psychedelic new psychoactive substances (nps) showing biased agonism at the 5-ht2ar through simultaneous use of beta-arrestin 2 and minigalphaq bioassays. Biochem. Pharmacol. 2020, 182, 114251. [Google Scholar] [CrossRef]
- Abiuso, A.M.B.; Varela, M.L.; Haro Durand, L.; Besio Moreno, M.; Marcos, A.; Ponzio, R.; Rivarola, M.A.; Belgorosky, A.; Pignataro, O.P.; Berensztein, E.; et al. Histamine h4 receptor as a novel therapeutic target for the treatment of leydig-cell tumours in prepubertal boys. Eur. J. Cancer 2018, 91, 125–135. [Google Scholar] [CrossRef]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. Nanoluc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Ayoub, M.A.; Landomiel, F.; Gallay, N.; Jegot, G.; Poupon, A.; Crepieux, P.; Reiter, E. Assessing gonadotropin receptor function by resonance energy transfer-based assays. Front. Endocrinol. (Lausanne) 2015, 6, 130. [Google Scholar] [CrossRef] [Green Version]
- Perkovska, S.; Mejean, C.; Ayoub, M.A.; Li, J.; Hemery, F.; Corbani, M.; Laguette, N.; Ventura, M.A.; Orcel, H.; Durroux, T.; et al. V1b vasopressin receptor trafficking and signaling: Role of arrestins, g proteins and src kinase. Traffic 2018, 19, 58–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copeland, R.A. The drug-target residence time model: A 10-year retrospective. Nat. Rev. Drug Discov. 2016, 15, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Castro, M.; Rinken, A.; Leurs, R.; Hill, S.J.; Vischer, H.F. Ligand residence time at g-protein-coupled receptors-why we should take our time to study it. Mol. Pharmacol. 2015, 88, 552–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Mocking, T.A.M.; Verweij, E.W.E.; Vischer, H.F.; Leurs, R. Homogeneous, real-time nanobret binding assays for the histamine h3 and h4 receptors on living cells. Mol. Pharmacol. 2018, 94, 1371–1381. [Google Scholar] [CrossRef] [Green Version]
- Bartole, E.; Littmann, T.; Tanaka, M.; Ozawa, T.; Buschauer, A.; Bernhardt, G. [(3)h]ur-deba176: A 2,4-diaminopyrimidine-type radioligand enabling binding studies at the human, mouse, and rat histamine h4 receptors. J. Med. Chem. 2019, 62, 8338–8356. [Google Scholar] [CrossRef]
- Schihada, H.; Ma, X.; Zabel, U.; Vischer, H.F.; Schulte, G.; Leurs, R.; Pockes, S.; Lohse, M.J. Development of a conformational histamine h3 receptor biosensor for the synchronous screening of agonists and inverse agonists. ACS Sens. 2020, 5, 1734–1742. [Google Scholar] [CrossRef]
- Schrage, R.; De Min, A.; Hochheiser, K.; Kostenis, E.; Mohr, K. Superagonism at g protein-coupled receptors and beyond. Br. J. Pharmacol. 2016, 173, 3018–3027. [Google Scholar] [CrossRef] [Green Version]
- Lieb, S.; Littmann, T.; Plank, N.; Felixberger, J.; Tanaka, M.; Schafer, T.; Krief, S.; Elz, S.; Friedland, K.; Bernhardt, G.; et al. Label-free versus conventional cellular assays: Functional investigations on the human histamine h1 receptor. Pharmacol. Res. 2016, 114, 13–26. [Google Scholar] [CrossRef]
- Strasser, A.; Wittmann, H.J.; Kunze, M.; Elz, S.; Seifert, R. Molecular basis for the selective interaction of synthetic agonists with the human histamine h1-receptor compared with the guinea pig h1-receptor. Mol. Pharmacol. 2009, 75, 454–465. [Google Scholar] [CrossRef] [Green Version]
- Costa, T.; Herz, A. Antagonists with negative intrinsic activity at delta opioid receptors coupled to gtp-binding proteins. Proc. Natl. Acad. Sci. USA 1989, 86, 7321–7325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smit, M.J.; Leurs, R.; Alewijnse, A.E.; Blauw, J.; Van Nieuw Amerongen, G.P.; Van De Vrede, Y.; Roovers, E.; Timmerman, H. Inverse agonism of histamine h2 antagonist accounts for upregulation of spontaneously active histamine h2 receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 6802–6807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakker, R.A.; Wieland, K.; Timmerman, H.; Leurs, R. Constitutive activity of the histamine h(1) receptor reveals inverse agonism of histamine h(1) receptor antagonists. Eur. J. Pharmacol. 2000, 387, R5–R7. [Google Scholar] [CrossRef]
- Alewijnse, A.E.; Smit, M.J.; Hoffmann, M.; Verzijl, D.; Timmerman, H.; Leurs, R. Constitutive activity and structural instability of the wild-type human h2 receptor. J. Neurochem. 1998, 71, 799–807. [Google Scholar] [CrossRef]
- Diaz Nebreda, A.; Zappia, C.D.; Rodriguez Gonzalez, A.; Sahores, A.; Sosa, M.; Burghi, V.; Monczor, F.; Davio, C.; Fernandez, N.; Shayo, C. Involvement of histamine h1 and h2 receptor inverse agonists in receptor’s crossregulation. Eur. J. Pharmacol. 2019, 847, 42–52. [Google Scholar] [CrossRef]
- Wifling, D.; Bernhardt, G.; Dove, S.; Buschauer, A. The extracellular loop 2 (ecl2) of the human histamine h4 receptor substantially contributes to ligand binding and constitutive activity. PLoS ONE 2015, 10, e0117185. [Google Scholar] [CrossRef] [Green Version]
- Seifert, R.; Wenzel-Seifert, K.; Burckstummer, T.; Pertz, H.H.; Schunack, W.; Dove, S.; Buschauer, A.; Elz, S. Multiple differences in agonist and antagonist pharmacology between human and guinea pig histamine h1-receptor. J. Pharmacol. Exp. Ther. 2003, 305, 1104–1115. [Google Scholar] [CrossRef] [Green Version]
- Igel, P.; Geyer, R.; Strasser, A.; Dove, S.; Seifert, R.; Buschauer, A. Synthesis and structure-activity relationships of cyanoguanidine-type and structurally related histamine h4 receptor agonists. J. Med. Chem. 2009, 52, 6297–6313. [Google Scholar] [CrossRef]
- Igel, P.; Schneider, E.; Schnell, D.; Elz, S.; Seifert, R.; Buschauer, A. N(g)-acylated imidazolylpropylguanidines as potent histamine h4 receptor agonists: Selectivity by variation of the n(g)-substituent. J. Med. Chem. 2009, 52, 2623–2627. [Google Scholar] [CrossRef]
- Appl, H.; Holzammer, T.; Dove, S.; Haen, E.; Strasser, A.; Seifert, R. Interactions of recombinant human histamine h(1)r, h(2)r, h(3)r, and h(4)r receptors with 34 antidepressants and antipsychotics. Naunyn Schmiedebergs. Arch. Pharmacol. 2012, 385, 145–170. [Google Scholar] [CrossRef]
- Bakker, R.A.; Schoonus, S.B.; Smit, M.J.; Timmerman, H.; Leurs, R. Histamine h(1)-receptor activation of nuclear factor-kappa b: Roles for g beta gamma- and g alpha(q/11)-subunits in constitutive and agonist-mediated signaling. Mol. Pharmacol. 2001, 60, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, P.; Erdmann, D.; Biselli, S.; Kagermeier, N.; Elz, S.; Bernhardt, G.; Buschauer, A. [(3) h]ur-de257: Development of a tritium-labeled squaramide-type selective histamine h2 receptor antagonist. ChemMedChem 2015, 10, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Leurs, R.; Smit, M.J.; Menge, W.M.; Timmerman, H. Pharmacological characterization of the human histamine h2 receptor stably expressed in chinese hamster ovary cells. Br. J. Pharmacol. 1994, 112, 847–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.D.; van Rijn, R.M.; Ling, P.; Bakker, R.A.; Thurmond, R.L.; Leurs, R. Evaluation of histamine h1-, h2-, and h3-receptor ligands at the human histamine h4 receptor: Identification of 4-methylhistamine as the first potent and selective h4 receptor agonist. J. Pharmacol. Exp. Ther. 2005, 314, 1310–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.D.; Smits, R.A.; Bakker, R.A.; van Dam, C.M.; de Esch, I.J.; Leurs, R. Discovery of s-(2-guanidylethyl)-isothiourea (vuf 8430) as a potent nonimidazole histamine h4 receptor agonist. J. Med. Chem. 2006, 49, 6650–6651. [Google Scholar] [CrossRef] [PubMed]
- Ligneau, X.; Perrin, D.; Landais, L.; Camelin, J.C.; Calmels, T.P.; Berrebi-Bertrand, I.; Lecomte, J.M.; Parmentier, R.; Anaclet, C.; Lin, J.S.; et al. Bf2.649 [1-{3-[3-(4-chlorophenyl)propoxy]propyl}piperidine, hydrochloride], a nonimidazole inverse agonist/antagonist at the human histamine h3 receptor: Preclinical pharmacology. J. Pharmacol. Exp. Ther. 2007, 320, 365–375. [Google Scholar] [CrossRef]
- Igel, P.; Schnell, D.; Bernhardt, G.; Seifert, R.; Buschauer, A. Tritium-labeled n1-[3-(1h-imidazol-4-yl)propyl]-n2-propionylguanidine ([3h]ur-pi294), a high-affinity histamine h3 and h4 receptor radioligand. ChemMedChem 2009, 4, 225–231. [Google Scholar] [CrossRef]
- Thurmond, R.L.; Desai, P.J.; Dunford, P.J.; Fung-Leung, W.P.; Hofstra, C.L.; Jiang, W.; Nguyen, S.; Riley, J.P.; Sun, S.; Williams, K.N.; et al. A potent and selective histamine h4 receptor antagonist with anti-inflammatory properties. J. Pharmacol. Exp. Ther. 2004, 309, 404–413. [Google Scholar] [CrossRef] [Green Version]
- Brunskole, I.; Strasser, A.; Seifert, R.; Buschauer, A. Role of the second and third extracellular loops of the histamine h(4) receptor in receptor activation. Naunyn Schmiedebergs. Arch. Pharmacol. 2011, 384, 301–317. [Google Scholar] [CrossRef]
- Schneider, E.H.; Schnell, D.; Papa, D.; Seifert, R. High constitutive activity and a g-protein-independent high-affinity state of the human histamine h4-receptor. Biochemistry 2009, 48, 1424–1438. [Google Scholar] [CrossRef]
- Nordemann, U.; Wifling, D.; Schnell, D.; Bernhardt, G.; Stark, H.; Seifert, R.; Buschauer, A. Luciferase reporter gene assay on human, murine and rat histamine h4 receptor orthologs: Correlations and discrepancies between distal and proximal readouts. PLoS ONE 2013, 8, e73961. [Google Scholar] [CrossRef] [PubMed]
- Cowart, M.D.; Altenbach, R.J.; Liu, H.; Hsieh, G.C.; Drizin, I.; Milicic, I.; Miller, T.R.; Witte, D.G.; Wishart, N.; Fix-Stenzel, S.R.; et al. Rotationally constrained 2,4-diamino-5,6-disubstituted pyrimidines: A new class of histamine h4 receptor antagonists with improved druglikeness and in vivo efficacy in pain and inflammation models. J. Med. Chem. 2008, 51, 6547–6557. [Google Scholar] [CrossRef] [PubMed]
- Tolkovsky, A.M.; Levitzki, A. Mode of coupling between the beta-adrenergic receptor and adenylate cyclase in turkey erythrocytes. Biochemistry 1978, 17, 3795. [Google Scholar] [CrossRef] [PubMed]
- Okashah, N.; Wan, Q.; Ghosh, S.; Sandhu, M.; Inoue, A.; Vaidehi, N.; Lambert, N.A. Variable g protein determinants of gpcr coupling selectivity. Proc. Natl. Acad. Sci. USA 2019, 116, 12054–12059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilyaskina, O.S.; Lemoine, H.; Bünemann, M. Lifetime of muscarinic receptor–g-protein complexes determines coupling efficiency and g-protein subtype selectivity. Proc. Natl. Acad. Sci. USA 2018, 115, 5016–5021. [Google Scholar] [CrossRef] [Green Version]
- Samama, P.; Cotecchia, S.; Costa, T.; Lefkowitz, R.J. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J. Biol. Chem. 1993, 268, 4625–4636. [Google Scholar]
- Kenakin, T. Efficacy as a vector: The relative prevalence and paucity of inverse agonism. Mol. Pharmacol. 2004, 65, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Elz, S.; Kramer, K.; Pertz, H.H.; Detert, H.; Ter Laak, A.M.; Kuhne, R.; Schunack, W. Histaprodifens: Synthesis, pharmacological in vitro evaluation, and molecular modeling of a new class of highly active and selective histamine h(1)-receptor agonists. J. Med. Chem. 2000, 43, 1071–1084. [Google Scholar] [CrossRef]
- Durant, G.J.; Ganellin, C.R.; Hills, D.W.; Miles, P.D.; Parsons, M.E.; Pepper, E.S.; White, G.R. The histamine h2-receptor agonist impromidine: Synthesis and structure activity considerations. J. Med. Chem. 1985, 28, 1414–1422. [Google Scholar] [CrossRef]
- Jablonowski, J.A.; Grice, C.A.; Chai, W.; Dvorak, C.A.; Venable, J.D.; Kwok, A.K.; Ly, K.S.; Wei, J.; Baker, S.M.; Desai, P.J.; et al. The first potent and selective non-imidazole human histamine h4 receptor antagonists. J. Med. Chem. 2003, 46, 3957–3960. [Google Scholar] [CrossRef]
- Littmann, T.; Buschauer, A.; Bernhardt, G. Split luciferase-based assay for simultaneous analyses of the ligand concentration- and time-dependent recruitment of beta-arrestin2. Anal. Biochem. 2019, 573, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Prusoff, W.H. Relationship between the inhibition constant (k1) and the concentration of inhibitor which causes 50 per cent inhibition (i50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [PubMed]
- Kagermeier, N.; Werner, K.; Keller, M.; Baumeister, P.; Bernhardt, G.; Seifert, R.; Buschauer, A. Dimeric carbamoylguanidine-type histamine h2 receptor ligands: A new class of potent and selective agonists. Bioorg. Med. Chem. 2015, 23, 3957–3969. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Höring, C.; Seibel, U.; Tropmann, K.; Grätz, L.; Mönnich, D.; Pitzl, S.; Bernhardt, G.; Pockes, S.; Strasser, A. A Dynamic, Split-Luciferase-Based Mini-G Protein Sensor to Functionally Characterize Ligands at All Four Histamine Receptor Subtypes. Int. J. Mol. Sci. 2020, 21, 8440. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228440
Höring C, Seibel U, Tropmann K, Grätz L, Mönnich D, Pitzl S, Bernhardt G, Pockes S, Strasser A. A Dynamic, Split-Luciferase-Based Mini-G Protein Sensor to Functionally Characterize Ligands at All Four Histamine Receptor Subtypes. International Journal of Molecular Sciences. 2020; 21(22):8440. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228440
Chicago/Turabian StyleHöring, Carina, Ulla Seibel, Katharina Tropmann, Lukas Grätz, Denise Mönnich, Sebastian Pitzl, Günther Bernhardt, Steffen Pockes, and Andrea Strasser. 2020. "A Dynamic, Split-Luciferase-Based Mini-G Protein Sensor to Functionally Characterize Ligands at All Four Histamine Receptor Subtypes" International Journal of Molecular Sciences 21, no. 22: 8440. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228440