mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors
Abstract
:1. Introduction
2. Results
2.1. MYC and E2F Gene Expression Is Connected with Tumor Type and Grade
2.2. miRNA Expression Depends on Tumor’s Histopathology and WHO Grade
2.3. Relationship between Gene Expression of Genes from MYC and E2F Families and miRNAs
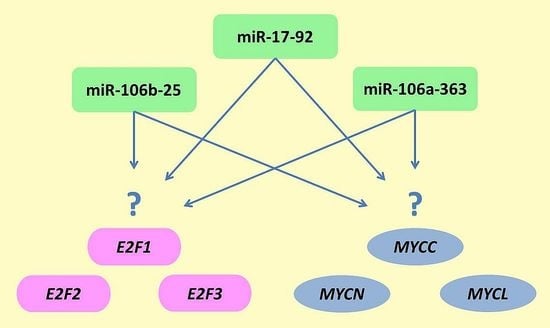
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Samples
4.2. RNA Isolation and Reverse Transcription
4.3. Reverse Transcription and Quantification of Gene Expression by qRT-PCR
4.4. Reverse Transcription and Detection of miRNA Expression by qRT-PCR
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EP | Ependymoma |
| MB | Medulloblastoma |
| PA | Pilocytic astrocytoma |
References
- Visani, M.; de Biase, D.; Marucci, G.; Cerasoli, S.; Nigrisoli, E.; Bacchi Reggiani, M.L.; Albani, F.; Baruzzi, A.; Pession, A. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol. Oncol. 2014, 8, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T. miRiad Roles for the miR-17-92 Cluster in Development and Disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Aguda, B.D.; Kim, Y.; Piper-Hunter, M.G.; Friedman, A.; Marsh, C.B. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. USA 2008, 105, 19678–19683. [Google Scholar] [CrossRef] [Green Version]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a Autoregulatory Feedback Loop. J. Biol. Chem. 2006, 282, 2135–2143. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, D.; Govindaraj, V.; Kar, S. Alteration in microRNA-17-92 dynamics accounts for differential nature of cellular proliferation. FEBS Lett. 2018, 592, 446–458. [Google Scholar] [CrossRef]
- Wang, G.; Yang, Z. Stability and Hopf Bifurcation Analysis in a Delayed Myc/E2F/miR-17-92 Network Involving Interlinked Positive and Negative Feedback Loops. Discret. Dyn. Nat. Soc. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med. Sci. 2018, 15, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Moi, L.; Braaten, T.; Al-Shibli, K.; Lund, E.; Busund, L.-T.R. Differential expression of the miR-17-92 cluster and miR-17 family in breast cancer according to tumor type; results from the Norwegian Women and Cancer (NOWAC) study. J. Transl. Med. 2019, 17, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Knudsen, K.N.; Nielsen, B.S.; Lindebjerg, J.; Hansen, T.F.; Holst, R.; Sørensen, F.B. microRNA-17 Is the Most Up-Regulated Member of the miR-17-92 Cluster during Early Colon Cancer Evolution. PLoS ONE 2015, 10, e0140503. [Google Scholar] [CrossRef]
- Tan, W.; Li, Y.; Lim, S.G.; Tan, T.M. miR-106b-25/miR-17-92 clusters: Polycistrons with oncogenic roles in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 5962–5972. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Miao, Y.; Xiao, B.; Huan, R.; Jiang, Z.; Meng, D.; Wang, Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J. Gastroenterol. Hepatol. 2009, 24, 652–657. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.C.; Wang, J.; Kong, J.; Qi, Y.; Quigg, R.J.; Li, X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. USA 2008, 105, 2889–2894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carraro, G.; El-Hashash, A.; Guidolin, D.; Tiozzo, C.; Turcatel, G.; Young, B.M.; De Langhe, S.P.; Bellusci, S.; Shi, W.; Parnigotto, P.P.; et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev. Biol. 2009, 333, 238–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez, Y.; Fernández-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [Green Version]
- Hong, L.; Lai, M.; Chen, M.; Xie, C.; Liao, R.; Kang, Y.J.; Xiao, C.; Hu, W.-Y.; Han, J.; Sun, P. The miR-17-92 Cluster of MicroRNAs Confers Tumorigenicity by Inhibiting Oncogene-Induced Senescence. Cancer Res. 2010, 70, 8547–8557. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Huang, Z.-P.; Seok, H.Y.; Ding, J.; Kataoka, M.; Zhang, Z.; Hu, X.; Wang, G.; Lin, Z.; Wang, S.; et al. mir-17–92 Cluster Is Required for and Sufficient to Induce Cardiomyocyte Proliferation in Postnatal and Adult Hearts. Circ. Res. 2013, 112, 1557–1566. [Google Scholar] [CrossRef]
- Bracken, A.P.; Ciro, M.; Cocito, A.; Helin, K. E2F target genes: Unraveling the biology. Trends Biochem. Sci. 2004, 29, 409–417. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nat. Cell Biol. 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, H.; Takeuchi, T.; Nishikawa, E.; Yanagisawa, K.; Hayashita, Y.; Ebi, H.; Yamada, H.; Suzuki, M.; Nagino, M.; Nimura, Y.; et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 2007, 26, 6099–6105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, J.; Shen, J.; Si, W.; Du, C.; Chen, D.; Xu, L.; Yao, M.; Fu, P.; Fan, W. Resveratrol promotes MICA/B expression and natural killer cell lysis of breast cancer cells by suppressing c-Myc/miR-17 pathway. Oncotarget 2017, 8, 65743–65758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Bian, C.; Liao, L.; Li, J.; Zhao, R.C. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res. Treat. 2010, 126, 565–575. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, S.; Zhang, Z.; He, J.; Xu, Y.; Liu, S. ROCK has a crucial role in regulating prostate tumor growth through interaction with c-Myc. Oncogene 2013, 33, 5582–5591. [Google Scholar] [CrossRef] [Green Version]
- Venturini, L.; Battmer, K.; Castoldi, M.; Schultheis, B.; Hochhaus, A.; Muckenthaler, M.U.; Ganser, A.; Eder, M.; Scherr, M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood 2007, 109, 4399–4405. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Trimarchi, J.M.; Fairchild, B.; Wen, J.; Lees, J.A. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1519–1524. [Google Scholar] [CrossRef]
- Ruiz-Pérez, M.V.; Henley, A.B.; Arsenian-Henriksson, M. The MYCN Protein in Health and Disease. Genes 2017, 8, 113. [Google Scholar] [CrossRef]
- Beltran, H. The N-myc Oncogene: Maximizing its Targets, Regulation, and Therapeutic Potential. Mol. Cancer Res. 2014, 12, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Yao, G.; Lee, T.J.; Mori, S.; Nevins, J.R.; You, L. A bistable Rb–E2F switch underlies the restriction point. Nat. Cell Biol. 2008, 10, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.V.; Johnson, D.G.; Jiang, H.; Xu, J.; Alonso, M.M.; Aldape, K.D.; Fuller, G.N.; Bekele, B.N.; Yung, W.K.A.; Gomez-Manzano, C.; et al. Transgenic E2F1 Expression in the Mouse Brain Induces a Human-Like Bimodal Pattern of Tumors. Cancer Res. 2007, 67, 4005–4009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirks, P.B.; Rutka, J.T.; Hubbard, S.L.; Mondal, S.; Hamel, P.A. The E2F-family proteins induce distinct cell cycle regulatory factors in p16-arrested, U343 astrocytoma cells. Oncogene 1998, 17, 867–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, T.G.; Grasfeder, L.L.; Carroll, A.L.; Kaiser, C.; Gillingham, C.L.; Lin, S.M.; Wickramasinghe, R.; Scott, M.P.; Wechsler-Reya, R.J. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 2003, 100, 7331–7336. [Google Scholar] [CrossRef] [Green Version]
- Swartling, F.J.; Grimmer, M.R.; Hackett, C.S.; Northcott, P.A.; Fan, Q.-W.; Goldenberg, D.D.; Lau, J.; Masic, S.; Nguyen, K.; Yakovenko, S.; et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010, 24, 1059–1072. [Google Scholar] [CrossRef] [Green Version]
- Skowron, P.; Ramaswamy, V.; Taylor, M.D. Genetic and molecular alterations across medulloblastoma subgroups. J. Mol. Med. 2015, 93, 1075–1084. [Google Scholar] [CrossRef] [Green Version]
- Knoepfler, P.S.; Cheng, P.F.; Eisenman, R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002, 16, 2699–2712. [Google Scholar] [CrossRef] [Green Version]
- Pomeroy, S.L.; Tamayo, P.; Gaasenbeek, M.; Sturla, L.M.; Angelo, M.; McLaughlin, M.E.; Kim, J.Y.; Goumnerova, L.C.; Black, P.M.; Lau, C.; et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002, 415, 436–442. [Google Scholar] [CrossRef]
- Higashi, M.; Sakai, K.; Fumino, S.; Aoi, S.; Furukawa, T.; Tajiri, T. The roles played by the MYCN, Trk, and ALK genes in neuroblastoma and neural development. Surg. Today 2019, 49, 721–727. [Google Scholar] [CrossRef]
- Swanson, A.A.; Raghunathan, A.; Jenkins, R.B.; Messing-Jünger, M.; Pietsch, T.; Clarke, M.J.; Kaufmann, T.J.; Giannini, C. Spinal Cord Ependymomas With MYCN Amplification Show Aggressive Clinical Behavior. J. Neuropathol. Exp. Neurol. 2019, 78, 791–797. [Google Scholar] [CrossRef]
- Malynn, B.A.; De Alboran, I.M.; O’Hagan, R.C.; Bronson, R.; Davidson, L.; Depinho, R.A.; Alt, F.W. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genome Res. 2000, 14, 1390–1399. [Google Scholar]
- Korshunov, A.; Schrimpf, D.; Ryzhova, M.; Sturm, D.; Chavez, L.; Hovestadt, V.; Sharma, T.; Habel, A.; Burford, A.; Jones, C.; et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017, 134, 507–516. [Google Scholar] [CrossRef] [PubMed]
- De Pontual, L.; Yao, E.; Callier, P.; Faivre, L.; Drouin, V.; Cariou, S.; Van Haeringen, A.; Geneviève, D.; Goldenberg, A.; Oufadem, M.; et al. Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011, 43, 1026–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, J.H.; Horn, S.; Otto, T.; Samans, B.; Heukamp, L.C.; Eilers, U.-C.; Krause, M.; Astrahantseff, K.; Klein-Hitpass, L.; Buettner, R.; et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer 2008, 122, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.; Thomson, J.M.; Hammond, S.M. Direct regulation of an oncogenic micro-RNA cluster by E2Ftranscription factors. J. Biol. Chem. 2007, 282, 2130–2134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, F.F.; Bischof, J.M.; Vanin, E.F.; Lulla, R.R.; Wang, M.; Sredni, S.T.; Rajaram, V.; Bonaldo, M.D.F.; Wang, D.; Goldman, S.; et al. Identification of MicroRNAs as Potential Prognostic Markers in Ependymoma. PLoS ONE 2011, 6, e25114. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. microRNA interaction with MAPK and AKT pathways in paediatric brain tumours—Preliminary results and review of the literature. Folia Neuropathol. 2020, 58, 123–132. [Google Scholar] [CrossRef]
- Lu, S.; Wang, S.; Geng, S.; Ma, S.; Liang, Z.; Jiao, B. Increased Expression of microRNA-17 Predicts Poor Prognosis in Human Glioma. J. Biomed. Biotechnol. 2012, 2012, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Chen, W.; Wang, J. MicroRNA-20a Regulates Glioma Cell Proliferation, Invasion, and Apoptosis by Targeting CUGBP Elav-Like Family Member 2. World Neurosurg. 2019, 121, e519–e527. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Fendler, W.; Zakrzewski, K.; Sikorska, B.; Grajkowska, W.; Dembowska-Bagińska, B.; Filipek, I.; Stefańczyk, Ł.; Liberski, P.P. Altered MicroRNA Expression Is Associated with Tumor Grade, Molecular Background and Outcome in Childhood Infratentorial Ependymoma. PLoS ONE 2016, 11, e0158464. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, R.; Chen, X.; Mu, Y.; Ai, J.; Shi, C.; Liu, Y.; Shi, C.; Sun, L.; Rainov, N.G.; et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J. Mol. Med. 2011, 89, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Z.; Chen, Z.; Lin, L.; Wang, Y.; Sailike, D.; Luo, K.; Du, G.; Xiang, X.; Jiafu, G.D. MicroRNA-106a-5p facilitates human glioblastoma cell proliferation and invasion by targeting adenomatosis polyposis coli protein. Biochem. Biophys. Res. Commun. 2016, 481, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Chen, X.; Wang, S.; Xia, X.; Shi, Y.; Guan, W.; Shao, N.; Qu, H.; Yang, C.; Zhang, Y.; et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur. J. Cancer. 2010, 46, 1640–1649. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.-I.; Hsu, C.-H.; Lee, K.-H.; Lin, J.-T.; Chen, C.-S.; Chang, K.-C.; Su, C.-Y.; Hsiao, M.; Lu, P.-J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. [Google Scholar] [CrossRef] [Green Version]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Wu, X.; Wang, Z.; Li, B.; Zhu, X. Effect of miR-18a overexpression on the radiosensitivity of non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 643–648. [Google Scholar]
- Tsujiura, M.; Komatsu, S.; Ichikawa, D.; Shiozaki, A.; Konishi, H.; Takeshita, H.; Moriumura, R.; Nagata, H.; Kawaguchi, T.; Hirajima, S.; et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2014, 18, 271–279. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, M.; Deng, D. c-Fos/microRNA-18a feedback loop modulates the tumor growth via HMBOX1 in human gliomas. Biomed. Pharmacother. 2018, 107, 1705–1711. [Google Scholar] [CrossRef]
- Song, Y.; Wang, P.; Zhao, W.; Yao, Y.; Liu, X.; Ma, J.; Xue, Y.; Liu, Y. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 2014, 324, 54–64. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Liu, J.; Li, L.; Yang, X.; Zhao, J.; Wu, M.; Ye, M. Upregulation of MicroRNA 18b Contributes to the Development of Colorectal Cancer by Inhibiting CDKN2B. Mol. Cell. Biol. 2017, 37, e00391-17. [Google Scholar] [CrossRef] [Green Version]
- Fonseca-Sanchéz, M.A.; Pérez-Plasencia, C.; Fernández-Retana, J.; Arechaga-Ocampo, E.; Marchat, L.A.; Rodríguez-Cuevas, S.; Bautista-Piña, V.; Arellano-Anaya, Z.E.; Flores-Pérez, A.; Diaz-Chávez, J.; et al. microRNA-18b is upregulated in breast cancer and modulates genes involved in cell migration. Oncol. Rep. 2013, 30, 2399–2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conti, A.; Romeo, S.G.; Cama, A.; La Torre, D.; Barresi, V.; Pezzino, G.; Tomasello, C.; Cardali, S.; Angileri, F.F.; Polito, F.; et al. MiRNA expression profiling in human gliomas: Upregulated miR-363 increases cell survival and proliferation. Tumor Biol. 2016, 37, 14035–14048. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, M.; Grochowski, C.; Litak, J.; Osuchowska, I.; Maciejewski, R.; Kamieniak, P. Recent Trends of microRNA Significance in Pediatric Population Glioblastoma and Current Knowledge of Micro RNA Function in Glioblastoma Multiforme. Int. J. Mol. Sci. 2020, 21, 3046. [Google Scholar] [CrossRef]
- El Baroudi, M.; Corà, D.; Bosia, C.; Osella, M.; Caselle, M. A Curated Database of miRNA Mediated Feed-Forward Loops Involving MYC as Master Regulator. PLoS ONE 2011, 6, e14742. [Google Scholar] [CrossRef]




| Gene | MB vs. PA | MB vs. EP | EP vs. PA |
|---|---|---|---|
| MYCC | 0.788019 | 0.471420 | 0.164877 |
| MYCN | 0.012457 | 0.375058 | 0.535525 |
| MYCL | 0.011674 | 0.000109 | 0.044045 |
| E2F1 | 0.000000 | 0.013521 | 0.014328 |
| E2F2 | 0.000000 | 0.000023 | 1.000000 |
| E2F3 | 0.132115 | 0.010907 | 1.000000 |
| miRNA | MB vs. PA | MB vs. EP | EP vs. PA | |
|---|---|---|---|---|
| 17-92 | miR-17-5p | 0.0000,66 | 1.0000,00 | 0.0018,81 |
| miR-18a | 0.0112,56 | 0.2688,29 | 0.6894,59 | |
| miR-19a | 0.0002,08 | 0.2688,29 | 0.6894,59 | |
| miR-20a | 0.0000,10 | 0.7671,39 | 0.0013,14 | |
| miR-19b | 0.0001,28 | 0.0765,68 | 0.0343,83 | |
| miR-92 | 0.0030,20 | 1.0000,00 | 0.0288,40 | |
| 106b-25 | miR-106b | 0.0001,07 | 0.1327,58 | 0.0001,70 |
| miR-93 | 0.0001,07 | 0.0076,12 | 0.0003,19 | |
| miR-25 | 0.0001,07 | 0.0526,61 | 0.0002,33 | |
| 106a-363 | miR-106a | 0.0001,07 | 0.0372,52 | 0.0042,90 |
| miR-18b | 0.0083,77 | 0.1771,93 | 0.8113,97 | |
| miR-19b-2 A | 0.0001,28 | 0.0765,68 | 0.0343,83 | |
| miR-20b | 0.0837,45 | 0.3829,15 | 0.6841,70 | |
| miR-92-2 B | 0.0030,20 | 1.0000,00 | 0.0288,40 | |
| miR-363 | 0.0000,00 | 0.0000,00 | 1.0000,00 |
| Gene–Gene | MB | EP | PA |
|---|---|---|---|
| MYCC-E2F1 | −0.21 | 0.55 | 0.75 |
| MYCC-E2F2 | −0.13 | 0.30 | 0.61 |
| MYCC-E2F3 | 0.19 | 0.54 | 0.72 |
| MYCN-E2F1 | 0.40 | 0.66 | 0.81 |
| MYCN-E2F2 | 0.51 | 0.60 | 0.79 |
| MYCN-E2F3 | −0.21 | 0.55 | 0.79 |
| MYCL-E2F1 | 0.21 | 0.25 | 0.37 |
| MYCL-E2F2 | 0.38 | 0.29 | 0.55 |
| MYCL-E2F3 | −0.22 | 0.62 | 0.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors. Int. J. Mol. Sci. 2021, 22, 543. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020543
Gruszka R, Zakrzewski K, Liberski PP, Zakrzewska M. mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors. International Journal of Molecular Sciences. 2021; 22(2):543. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020543
Chicago/Turabian StyleGruszka, Renata, Krzysztof Zakrzewski, Paweł Piotr Liberski, and Magdalena Zakrzewska. 2021. "mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors" International Journal of Molecular Sciences 22, no. 2: 543. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020543