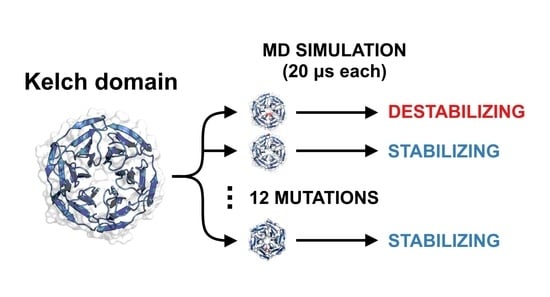
KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability
Abstract
:1. Introduction
2. Methodology
2.1. System Setup
2.2. Simulation Protocol
2.3. Analyses
3. Results
3.1. Mutations Have Differential Effects on Kelch’s -Sheets
3.2. Stabilizing and Destabilizing Mutations Both Result in Global Structural Deformation
3.3. Destabilizing Mutations Shift the Backbone Dihedrals of Kelch
3.4. Loss of Hydrogen Bonds Due to Destabilizing Mutations
3.5. Limited Fluctuations at Both the ANCHOR-Mutant Mutation Site and NRF2 Binding Site
4. Discussion
4.1. Structural Mutation Prediction
4.2. -Blades I, II and III Are Less Stable Than Blades IV, V and VI
4.3. ANCHOR Mutant
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dill, K.A.; MacCallum, J.L. The Protein-Folding Problem, 50 Years On. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Tunyasuvunakool, K.; Ronneberger, O.; Bates, R.; Žídek, A.; Bridgland, A.; et al. AlphaFold 2. Available online: https://predictioncenter.org/casp14/doc/presentations/2020_12_01_TS_predictor_AlphaFold2.pdf (accessed on 17 May 2021).
- Service, R.F. ‘The game has changed.’ AI triumphs at protein folding. Science 2020, 370, 1144–1145. [Google Scholar] [CrossRef] [PubMed]
- White, M.B.; Amos, J.; Hsu, J.M.C.; Gerrard, B.; Finn, P.; Dean, M. A frame-shift mutation in the cystic fibrosis gene. Nature 1990, 344, 665–667. [Google Scholar] [CrossRef]
- Moore, D.J.; Zhang, L.; Dawson, T.M.; Dawson, V.L. A missense mutation (L166P) in DJ-1, linked to familial Parkinsons disease, confers reduced protein stability and impairs homo-oligomerization. J. Neurochem. 2013, 87, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Lyst, M.J.; Bird, A. Rett syndrome: A complex disorder with simple roots. Nat. Rev. Genet. 2015, 16, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Minde, D.P.; Anvarian, Z.; Rüdiger, S.G.D.; Maurice, M.M. Messing up disorder: How do missense mutations in the tumor suppressor protein APC lead to cancer? Mol. Cancer 2011, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redler, R.L.; Das, J.; Diaz, J.R.; Dokholyan, N.V. Protein Destabilization as a Common Factor in Diverse Inherited Disorders. J. Mol. Evol. 2016, 82, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Tokuriki, N.; Tawfik, D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol 2009, 19, 596–604. [Google Scholar] [CrossRef]
- Engin, H.B.; Kreisberg, J.F.; Carter, H. Structure-Based Analysis Reveals Cancer Missense Mutations Target Protein Interaction Interfaces. PLoS ONE 2016, 11, e0152929. [Google Scholar] [CrossRef]
- Ozdemeir, E.S.; Gursoy, A.; Keskin, O. Analysis of Single Amino Acid Variations in Singlet Hot Spots of Protein-Protein Interfaces. Bioinformatics 2018, 34, 795–801. [Google Scholar] [CrossRef]
- Brown, D.K.; Bishop, Ö.T. Role of Structural Bioinformatics in Drug Discovery by Computational SNP Analysis. Glob. Heart 2017, 12, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, S.; Chaudhary, V.; Goswami, G.K.; Mathur, N. Computational approaches for predicting mutant protein stability. J. Comput. Aided Mol. Des. 2016, 30, 401–412. [Google Scholar] [CrossRef]
- Lee, A.C.L.; Harris, J.L.; Khanna, K.K.; Hong, J.H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niroula, A.; Vihinen, M. Variation Interpretation Predictors: Principles, Types, Performance, and Choice. Hum. Mutat. 2016, 37, 579–597. [Google Scholar] [CrossRef]
- Pasupati, S.; Doss, C.G.P. Molecular Dynamics: New Frontier in Personalized Medicine. In Personalized Medicine; Donev, R., Ed.; Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2016; Chapter 7; Volume 102, pp. 181–224. [Google Scholar] [CrossRef]
- Rost, B.; Radivojac, P.; Bromberg, Y. Protein function in precision medicine: Deep understanding with machine learning. FEBS Lett. 2016, 590, 2327–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahlgren, C.; Meinander, A.; Zhang, H.; Cheng, F.; Preis, M.; Xu, C.; Salminen, T.A.; Toivola, D.; Abankwa, D.; Rosling, A.; et al. Tailored Approaches in Drug Development and Diagnostics: From Molecular Design to Biological Model Systems. Adv. Healthc. Mater. 2017, 6, 1700258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanavia, T.; Birolo, G.; Montanucci, L.; Turina, P.; Capriotti, E.; Fariselli, P. Limitations and challenges in protein stability prediction upon genome variations: Towards future applications in precision medicine. Comput. Struct. Biotechnol. J. 2020, 18, 1968–1979. [Google Scholar] [CrossRef]
- Geng, C.; Xue, L.C.; Roel-Touris, J.; Bonvin, A.M.J.J. Finding the ΔΔG spot: Are predictors of binding affinity changes upon mutations in protein–protein interactions ready for it? Wiley Interdiscip. Rev. Comput. Mol. Sci. 2019, 9, e1410. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [Green Version]
- Fariselli, P.; Martelli, P.L.; Savojardo, C.; Casadio, R. INPS: Predicting the impact of non-synonymous variations on protein stability from sequence. Bioinformatics 2015, 31, 2816–2821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, L.; Stantic, B.; Sattar, A.; Zhou, Y. EASE-MM: Sequence-Based Prediction of Mutation-Induced Stability Changes with Feature-Based Multiple Models. J. Mol. Biol. 2016, 428, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A Server for Predicting Effects of Mutations on Protein Stability. Nucleic Acids Res. 2017, 45, 229–235. [Google Scholar] [CrossRef] [Green Version]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yves, D.; Kwasigroch, J.M.; Gilis, D.; Rooman, M. PoPMuSiC 2.1: A web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinform. 2011, 12. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Leaver-Fay, A.; Baker, D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 2011, 79, 830–838. [Google Scholar] [CrossRef] [Green Version]
- Rohl, C.A.; Strauss, C.E.; Misura, K.M.; Baker, D. Protein Structure Prediction Using Rosetta. In Numerical Computer Methods, Part D; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2004; Volume 383, pp. 66–93. [Google Scholar] [CrossRef]
- Frappier, V.; Chariter, M.; Najmanovich, R.J. ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015, 43, W395–W400. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J. Chem. Inf. Model. 2019, 59, 1508–1514. [Google Scholar] [CrossRef]
- Dehghanpoor, R.; Ricks, E.; Hursh, K.; Gunderson, S.; Farhoodi, R.; Haspel, N.; Hutchinson, B.; Jagodzinski, F. Predicting the Effect of Single and Multiple Mutations on Protein Structural Stability. Molecules 2018, 23, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Huang, J.; Zhang, R.; Wang, F.; Zhang, Q.; Li, G.; Yan, J.; Zhang, H.; Yan, Y.; Xu, L. Convolution Neural Network-Based Prediction of Protein Thermostability. J. Chem. Inf. Model. 2019, 59, 4833–4843. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016, 32, 2936–2946. [Google Scholar] [CrossRef] [Green Version]
- Broom, A.; Jacobi, Z.; Trainor, K.; Meiering, E.M. Computational tools help improve protein stability but with a solubility tradeoff. J. Biol. Chem. 2017, 292, 14349–14361. [Google Scholar] [CrossRef] [Green Version]
- Fang, J. A critical review of five machine learning-based algorithms for predicting protein stability changes upon mutation. Brief. Bioinform. 2020, 21, 1285–1292. [Google Scholar] [CrossRef] [Green Version]
- Pucci, F.; Bernaerts, K.V.; Kwasigroch, J.M.; Rooman, M. Quantification of Biases in Predictions of Protein Stability Changes Upon Mutations. Bioinformatics 2018, 34, 3659–3665. [Google Scholar] [CrossRef] [Green Version]
- Usmanova, D.R.; Bogatyreva, N.S.; Bernard, J.A.; Eremina, A.A.; Gorshkova, A.A.; Kanevskiy, G.M.; Lonishin, L.R.; Meister, A.V.; Yakupova, A.G.; Kondrashov, F.A.; et al. Self-consistency Test Reveals Systematic Bias in Programs for Prediction Change of Stability Upon Mutation. Bioinformatics 2018, 34, 3653–3658. [Google Scholar] [CrossRef]
- Walsh, I.; Pollastri, G.; Tosatto, S.C.E. Correct machine learning on protein sequences: A peer-reviewing perspective. Brief. Bioinform. 2015, 17, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Vihinen, M. Performance of Protein Stability Predictors. Hum. Mutat. 2010, 31, 675–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potapov, V.; Cohen, M.; Schreiber, G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng. Des. Sel. 2009, 22, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the Impact of Mutations on Protein Conformation, Flexibility and Stability. Nucleic Acids Res. 2012, 46, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Wilson, C.J.; Karunatilleke, N.C.; Moselhy, M.H.; Karttunen, M.; Choy, W.Y. Exploring the Conformational Landscape of the Neh4 and Neh5 Domains of Nrf2 Using Two Different Force Fields and Circular Dichroism. J. Chem. Theory Comput. 2021, 17, 3145–3156. [Google Scholar] [CrossRef] [PubMed]
- Childers, M.C.; Daggett, V. Insights from molecular dynamics simulations for computational protein design. Mol. Syst. Des. Eng. 2017, 2, 9–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grasso, G.; Morbiducci, U.; Massai, D.; Tuszynski, J.A.; Danani, A.; Deriu, M.A. Destabilizing the AXH Tetramer by Mutations: Mechanisms and Potential Antiaggregation Strategies. Biophys. J. 2018, 114, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Gur, M.; Blackburn, E.A.; Ning, J.; Narayan, V.; Ball, K.L.; Walkinshaw, M.D.; Erman, B. Molecular dynamics simulations of site point mutations in the TPR domain of cyclophilin 40 identify conformational states with distinct dynamic and enzymatic properties. J. Chem. Phys. 2018, 148, 145101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koukos, P.I.; Glykos, N.M. Folding Molecular Dynamics Simulations Accurately Predict the Effect of Mutations on the Stability and Structure of a Vammin-Derived Peptide. J. Phys. Chem. B 2014, 118, 10076–10084. [Google Scholar] [CrossRef]
- Ochoa, R.; Soler, M.A.; Laio, A.; Cossio, P. Assessing the capability of in silico mutation protocols for predicting the finite temperature conformation of amino acids. Phys. Chem. Chem. Phys. 2018, 20, 25901–25909. [Google Scholar] [CrossRef] [Green Version]
- Pirolli, D.; Sciandra, F.; Bozzi, M.; Giardina, B.; Brancaccio, A.; De Rosa, M.C. Insights from Molecular Dynamics Simulations: Structural Basis for the V567D Mutation-Induced Instability of Zebrafish Alpha-Dystroglycan and Comparison with the Murine Model. PLoS ONE 2014, 9, e103866. [Google Scholar] [CrossRef] [Green Version]
- Steinbrecher, T.; Zhu, C.; Wang, L.; Abel, R.; Negron, C.; Pearlman, D.; Feyfant, E.; Duan, J.; Sherman, W. Predicting the Effect of Amino Acid Single-Point Mutations on Protein Stability—Large-Scale Validation of MD-Based Relative Free Energy Calculations. J. Mol. Biol. 2017, 429, 948–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Orioli, S.; Ianeselli, A.; Spagnolli, G.; A Beccara, S.; Gershenson, A.; Faccioli, P.; Wintrode, P.L. All-Atom Simulations Reveal How Single-Point Mutations Promote Serpin Misfolding. Biophys. J. 2018, 114, 2083–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, M.; McCully, M.E. Molecular dynamics simulations suggest stabilizing mutations in a de novo designed α/β protein. Protein Eng. Des. Sel. 2020, 32, 317–329. [Google Scholar] [CrossRef]
- Daday, C.; Mateyka, L.M.; Gräter, F. How ARVC-Related Mutations Destabilize Desmoplakin: An MD Study. Biophys. J. 2019, 116, 831–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuyoshi, S.; Kometani, M.; Watanabe, Y.; Hiratsuka, M.; Yamaotsu, N.; Hirono, S.; Manabe, N.; Takahashi, O.; Oda, A. Molecular Dynamics Simulations to Investigate the Influences of Amino Acid Mutations on Protein Three-Dimensional Structures of Cytochrome P450 2D6.1, 2, 10, 14A, 51, and 62. PLoS ONE 2016, 4, e0152946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, L.B.; Gintner, L.P.; Park, S.; Snow, C.D. Discriminating between stabilizing and destabilizing protein design mutations via recombination and simulation. Protein Eng. Des. Sel. 2015, 28, 259–267. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wang, L.; Sun, X.; Zhao, X. Investigating the Impact of Asp181 Point Mutations on Interactions between PTP1B and Phosphotyrosine Substrate. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Ndagi, U.; Mhlongo, N.N.; Soliman, M.E. The impact of Thr91 mutation on c-Src resistance to UM-164: Molecular dynamics study revealed a new opportunity for drug design. Mol. BioSyst. 2017, 13, 1157–1171. [Google Scholar] [CrossRef]
- Petukh, M.; Li, M.; Alexov, E. Predicting Binding Free Energy Change Caused by Point Mutations with Knowledge-Modified MM/PBSA Method. PLoS Comput. Biol. 2015, 11, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Rallapalli, K.L.; Komor, A.C.; Paesani, F. Computer simulations explain mutation-induced effects on the DNA editing by adenine base editors. Sci. Adv. 2020, 6, eaaz2309. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, S.; Srivastava, G.; Sharma, A. Molecular dynamics simulation and free energy landscape methods in probing L215H, L217R and L225M βI-tubulin mutations causing paclitaxel resistance in cancer cells. Biochem. Biophys. Res. Commun. 2016, 476, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Milkovic, L.; Zarkovic, N.; Saso, L. Controversy About Pharmacological Modulation of Nrf2 for Cancer Therapy. Redox Biol. 2017, 12, 727–732. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, 941–947. [Google Scholar] [CrossRef] [Green Version]
- Hast, B.E.; Cloer, E.W.; Goldfarb, D.; Li, H.; Siesser, P.F.; Yan, F.; Walter, V.; Zheng, N.; Haynes, D.N.; Major, M.B. Cancer-Derived Mutations in KEAP1 Impair NRF2 Degradation but not Ubiquitination. Cancer Res. 2013, 74, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Cloer, E.W.; Siesser, P.F.; Cousins, E.M.; Goldfarb, D.; Mowrey, D.D.; Harrison, J.S.; Weir, S.J.; Dokholyan, N.V.; Major, M.B. p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol. Cell. Biol. 2018, 38, e00644-17. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Killoran, R.C.; Brickenden, A.; Fan, J.; Yang, D.; Choy, W.Y. Molecular Effects of Cancer-Associated Somatic Mutations on the Structural and Target Recognition Properties of Keap1. Biochem. J. 2015, 467, 141–151. [Google Scholar] [CrossRef]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinform. 2014, 15, 1–30. [Google Scholar] [CrossRef] [Green Version]
- PyMOL. The PyMOL Molecular Graphics System; Version 2.4.0; Schrödinger, Inc.: New York, NY, USA, 2020. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pàll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softw. X 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; Groot, B.L.d.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Wong-ekkabut, J.; Karttunen, M. The good, the bad and the user in soft matter simulations. Biochim. Biophys. Acta 2016, 1858, 2529–2538. [Google Scholar] [CrossRef]
- Wong-ekkabut, J.; Miettinen, M.S.; Dias, C.; Karttunen, M. Static charges cannot drive a continuous flow of water molecules through a carbon nanotube. Nat. Nanotechnol. 2010, 5, 555–557. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Pietrucci, F.; Laio, A. A Collective Variable for the Efficient Exploration of Protein Beta-Sheet Structures: Application to SH3 and GB1. J. Chem. Theory Comput. 2009, 5, 2197–2201. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Branduardi, D.; Bussi, G.; Camilloni, C.; Provasi, D.; Raiteri, P.; Donadio, D.; Marinelli, F.; Pietrucci, F.; Broglia, R.A.; et al. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009, 180, 1961–1972. [Google Scholar] [CrossRef] [Green Version]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef] [Green Version]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.; Ziolek, R.M.; Gazzarrini, E.; Owen, D.M.; Lorenz, C.D. On the interaction of hyaluronic acid with synovial fluid lipid membranes. Phys. Chem. Chem. Phys. 2019, 21, 9845–9857. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, M.; Alexov, E. DelPhiPKa web server: Predicting pKa of proteins, RNAs and DNAs. Bioinformatics 2015, 32, 614–615. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Alexov, E. pKa predictions for proteins, RNAs, and DNAs with the Gaussian dielectric function using DelPhi pKa. Proteins 2015, 83, 2186–2197. [Google Scholar] [CrossRef] [Green Version]
- Parzen, E. On Estimation of a Probability Density Function and Mode. Ann. Math. Stat. 1962, 33, 1065–1076. [Google Scholar] [CrossRef]
- Rosenblatt, M. Remarks on Some Nonparametric Estimates of a Density Function. Ann. Math. Stat. 1956, 27, 832–837. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, Thermodynamic, and Structural Characterizations of the Association between Nrf2-DLGex Degron and Keap1. Mol. Cell. Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Kai, E.L.J.; Lu, L. Investigating the stability of dengue virus envelope protein dimer using well-tempered metadynamics simulations. Proteins 2020, 88, 643–653. [Google Scholar] [CrossRef]
- Tsuber, V.; Kadamov, Y.; Brautigam, L.; Warpman Berglund, U.; Helleday, T. Mutations in Cancer Cause Gain of Cysteine, Histidine, and Tryptophan at the Expense of a Net Loss of Arginine on the Proteome Level. Biomolecules 2017, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuishi, Y.; Motohashi, H.; Yamamoto, M. The Keap1–Nrf2 system in cancers: Stress response and anabolic metabolism. Front. Oncol. 2012, 2, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017, 617, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Eggler, A.L.; Mesecar, A.D.; van Breemen, R.B. Modification of Keap1 Cysteine Residues by Sulforaphane. Chem. Res. Toxicol. 2011, 24, 515–521. [Google Scholar] [CrossRef] [Green Version]
- Holland, R.; Hawkins, A.E.; Eggler, A.L.; Mesecar, A.D.; Fabris, D.; Fishbein, J.C. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem. Res. Toxicol. 2008, 21, 2051–2060. [Google Scholar] [CrossRef] [Green Version]
- Fujii, S.; Sawa, T.; Ihara, H.; Tong, K.I.; Ida, T.; Okamoto, T.; Ahtesham, A.K.; Ishima, Y.; Motohashi, H.; Yamamoto, M.; et al. The critical role of nitric oxide signaling, via protein S-guanylation and nitrated cyclic GMP, in the antioxidant adaptive response. J. Biol. Chem. 2010, 285, 23970–23984. [Google Scholar] [CrossRef] [Green Version]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrov, D.; Margreitter, C.; Grandits, M.; Oostenbrink, C.; Zagrovic, B. A Systematic Framework for Molecular Dynamics Simulations of Protein Post-Translational Modifications. PLoS Comput. Biol. 2013, 9, e1003154. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]












| Mutant | Exp. | Sim. | PP2 | SIFT | PMC | SDM | DM | ECM | mCSM | MUT | DGG |
|---|---|---|---|---|---|---|---|---|---|---|---|
| G333C | D | D | 1.00 | 0.05 | / | −1.45 | −0.76 | 1.25 | −1.92 | −0.63 | −1.98 |
| G350S | S/N | S/N | 0.99 | 0.22 | / | −3.40 | −1.00 | 0.19 | −1.13 | −1.15 | −0.69 |
| G364C | S/N | S/N | 1.00 | 0.03 | / | −1.37 | 0.81 | 0.35 | −1.48 | −0.70 | −1.46 |
| G379D | D | D | 1.00 | 0.00 | / | −2.72 | −1.15 | 0.98 | −2.99 | −0.79 | −2.83 |
| R413L | D | D | 1.00 | 0.00 | −0.58 | 0.04 | 0.24 | −0.38 | −0.68 | −0.71 | −1.23 |
| R415G | S/N | S/N | 0.95 | 0.43 | / | 2.18 | −0.70 | −0.80 | −1.42 | −0.86 | −0.40 |
| A427V | S/N | I | 1.00 | 0.00 | −0.59 | 0.57 | 3.16 | 0.40 | 0.22 | −0.73 | 0.26 |
| G430C | D | D | 1.00 | 0.00 | / | −1.52 | −0.33 | 1.14 | −1.80 | −0.79 | −1.77 |
| R470C | S/N | S/N | 0.88 | 0.01 | / | −0.51 | −0.75 | −0.42 | −0.13 | −1.00 | −0.22 |
| R470H | S/N | S/N | 1.00 | 0.03 | / | −0.14 | −0.84 | −0.31 | −0.82 | −1.26 | −0.28 |
| R470S | S/N | S/N | 0.99 | 0.08 | / | −1.12 | −0.57 | −0.27 | −0.23 | −1.21 | −0.42 |
| G476R | D | D | 1.00 | 0.00 | / | −3.19 | −1.36 | 0.75 | −0.99 | −0.62 | −2.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, C.J.; Chang, M.; Karttunen, M.; Choy, W.-Y. KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability. Int. J. Mol. Sci. 2021, 22, 5408. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22105408
Wilson CJ, Chang M, Karttunen M, Choy W-Y. KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability. International Journal of Molecular Sciences. 2021; 22(10):5408. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22105408
Chicago/Turabian StyleWilson, Carter J., Megan Chang, Mikko Karttunen, and Wing-Yiu Choy. 2021. "KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability" International Journal of Molecular Sciences 22, no. 10: 5408. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22105408