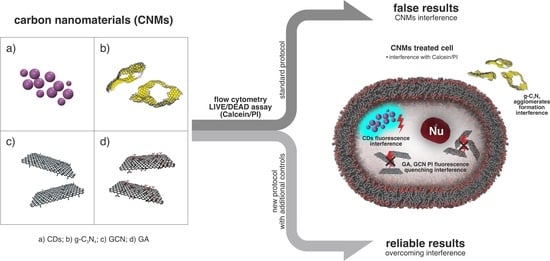
Toxicity of Carbon Nanomaterials—Towards Reliable Viability Assessment via New Approach in Flow Cytometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Properties of Materials
2.2. Optical Microscopy Imaging and MTT Interference
2.3. Interference of CNMs in Forward and Side Scatter Profiles in Flow Cytometry
2.4. Interference of CNMs in Spike-in Controls
2.5. Interference of QCDs in LIVE/DEAD Assay
2.6. Interference of g-C3N4 in LIVE/DEAD Assay
2.7. Interference of GA and GCN Samples in LIVE/DEAD Assay
3. Materials and Methods
3.1. Materials and Characterization
3.2. Cell Culture
3.3. Flow Cytometry Scatter Profiles and LIVE/DEAD Assay
3.4. Flow Cytometry Controls
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Wang, L.; Li, Y.; Feng, Y.; Feng, W. Carbon-based functional nanomaterials: Preparation, properties and applications. Compos. Sci. Technol. 2019, 179, 10–40. [Google Scholar] [CrossRef]
- Jaleel, J.A.; Pramod, K. Artful and multifaceted applications of carbon dot in biomedicine. J. Control. Release 2018, 269, 302–321. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Sakthivel, T.; Seal, S. Recent development in 2D materials beyond graphene. Prog. Mater. Sci. 2015, 73, 44–126. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.; Perrnan, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Du, D.; Lin, Y. Graphene and graphene-like 2D materials for optical biosensing and bioimaging: A review. 2D Mater. 2015, 2, 032004. [Google Scholar] [CrossRef]
- Bakandritsos, A.; Pykal, M.; Błoński, P.; Jakubec, P.; Chronopoulos, D.D.; Poláková, K.; Georgakilas, V.; Čépe, K.; Tomanec, O.; Ranc, V.; et al. Otyepka, Cyanographene and Graphene Acid: Emerging Derivatives Enabling High-Yield and Selective Functionalization of Graphene. ACS Nano 2017, 11, 2982–2991. [Google Scholar] [CrossRef]
- Liao, G.; He, F.; Li, Q.; Zhong, L.; Zhao, R.; Che, H.; Gao, H.; Fang, B. Emerging graphitic carbon nitride-based materials for biomedical applications. Prog. Mater. Sci. 2020, 112, 100666. [Google Scholar] [CrossRef]
- Hola, K.; Zhang, Y.; Wang, Y.; Giannelis, E.P.; Zboril, R.; Rogach, A.L. Carbon dots—Emerging light emitters for bioimaging, cancer therapy and optoelectronics. Nano Today 2014, 9, 590–603. [Google Scholar] [CrossRef]
- Wang, A.-J.; Li, H.; Huang, H.; Qian, Z.-S.; Feng, J.-J. Fluorescent graphene-like carbon nitrides: Synthesis, properties and applications. J. Mater. Chem. C 2016, 4, 8146–8160. [Google Scholar] [CrossRef]
- Devi, P.; Saini, S.; Kim, K.-H. The advanced role of carbon quantum dots in nanomedical applications. Biosens. Bioelectron. 2019, 141, 111158. [Google Scholar] [CrossRef]
- Malina, T.; Poláková, K.; Skopalík, J.; Milotová, V.; Hola, K.; Havrdová, M.; Tománková, K.B.; Čmiel, V.; Sefc, L.; Zbořil, R. Carbon dots for in vivo fluorescence imaging of adipose tissue-derived mesenchymal stromal cells. Carbon 2019, 152, 434–443. [Google Scholar] [CrossRef]
- Drasler, B.; Sayre, P.; Steinhäuser, K.G.; Fink, A.; Rothen-Rutishauser, B. In vitro approaches to assess the hazard of nanomaterials. NanoImpact 2017, 8, 99–116. [Google Scholar] [CrossRef]
- Krewski, D.; Acosta, D.; Andersen, M.; Anderson, H.; Bailar, J.C.; Boekelheide, K.; Brent, R.; Charnley, G.; Cheung, V.G.; Green, S.; et al. Toxicity Testing in the 21st Century: A Vision and a Strategy. J. Toxicol. Environ. Health Part B 2010, 13, 51–138. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.E.; Krewski, D. Toxicity Testing in the 21st Century: Bringing the Vision to Life. Toxicol. Sci. 2008, 107, 324–330. [Google Scholar] [CrossRef] [Green Version]
- Romeo, D.; Salieri, B.; Hischier, R.; Nowack, B.; Wick, P. An integrated pathway based on in vitro data for the human hazard assessment of nanomaterials. Environ. Int. 2020, 137, 105505. [Google Scholar] [CrossRef]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Interference of engineered nanoparticles with in vitro toxicity assays. Arch. Toxicol. 2012, 86, 1123–1136. [Google Scholar] [CrossRef]
- Ong, K.J.; MacCormack, T.; Clark, R.J.; Ede, J.D.; Ortega, V.A.; Felix, L.; Dang, M.K.M.; Ma, G.; Fenniri, H.; Veinot, J.G.C.; et al. Widespread Nanoparticle-Assay Interference: Implications for Nanotoxicity Testing. PLoS ONE 2014, 9, e90650. [Google Scholar] [CrossRef]
- Guadagnini, R.; Kenzaoui, B.H.; Walker, L.; Pojana, G.; Magdolenova, Z.; Bilanicova, D.; Saunders, M.; Juillerat-Jeanneret, L.; Marcomini, A.; Huk, A. Boland, Toxicity screenings of nanomaterials: Challenges due to interference with assay processes and components of classic in vitro tests. Nanotoxicology 2015, 9, 13–24. [Google Scholar] [CrossRef]
- Andraos, C.; Yu, I.J.; Gulumian, M. Interference: A Much-Neglected Aspect in High-Throughput Screening of Nanoparticles. Int. J. Toxicol. 2020, 39, 397–421. [Google Scholar] [CrossRef]
- Labouta, H.; Asgarian, N.; Rinker, K.; Cramb, D.T. Meta-Analysis of Nanoparticle Cytotoxicity via Data-Mining the Literature. ACS Nano 2019, 13, 1583–1594. [Google Scholar] [CrossRef]
- Wright, P.C.; Qin, H.; Choi, M.M.; Chiu, N.H.; Jia, Z. Carbon nanodots interference with lactate dehydrogenase assay in human monocyte THP-1 cells. SpringerPlus 2014, 3, 615. [Google Scholar] [CrossRef] [Green Version]
- Casey, A.; Herzog, E.; Davoren, M.; Lyng, F.; Byrne, H.; Chambers, G. Spectroscopic analysis confirms the interactions between single walled carbon nanotubes and various dyes commonly used to assess cytotoxicity. Carbon 2007, 45, 1425–1432. [Google Scholar] [CrossRef]
- Monteiro-Riviere, N.; Inman, A.O. Challenges for assessing carbon nanomaterial toxicity to the skin. Carbon 2006, 44, 1070–1078. [Google Scholar] [CrossRef]
- Wörle-Knirsch, J.M.; Pulskamp, A.K.; Krug, H.F.; Wörle-Knirsch, J.M.; Pulskamp, A.K.; Krug, H.F. Oops They Did It Again! Carbon Nanotubes Hoax Scientists in Viability Assays. Nano Lett. 2006, 6, 1261–1268. [Google Scholar] [CrossRef]
- Holder, A.; Goth-Goldstein, R.; Lucas, D.; Koshland, C.P. Particle-Induced Artifacts in the MTT and LDH Viability Assays. Chem. Res. Toxicol. 2012, 25, 1885–1892. [Google Scholar] [CrossRef] [Green Version]
- Belyanskaya, L.; Manser, P.; Spohn, P.; Bruinink, A.; Wick, P. The reliability and limits of the MTT reduction assay for carbon nanotubes-cell interaction. Carbon 2007, 45, 2643–2648. [Google Scholar] [CrossRef]
- Tuchin, V.V.; Tárnok, A.; Zharov, V.P. In vivo flow cytometry: A horizon of opportunities. Cytom. Part A 2011, 79, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Bakke, A.C. The Principles of Flow Cytometry. Lab. Med. 2001, 32, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Bohmer, N.; Rippl, A.; May, S.; Walter, A.; Heo, M.B.; Kwak, M.; Roesslein, M.; Song, N.W.; Wick, P.; Hirsch, C. Interference of engineered nanomaterials in flow cytometry: A case study. Colloids Surf. B: Biointerfaces 2018, 172, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, L.; Praus, P.; de Lima, M.J.B.P.; Sampaio, M.J.; Matýsek, D.; Ritz, M.; Dvorský, R.; Faria, J.L.; Silva, C.G. Graphitic carbon nitride nanosheets as highly efficient photocatalysts for phenol degradation under high-power visible LED irradiation. Mater. Res. Bull. 2018, 100, 322–332. [Google Scholar] [CrossRef]
- Svoboda, L.; Škuta, R.; Matějka, V.; Dvorský, R.; Matýsek, D.; Henych, J.; Mančík, P.; Praus, P. Graphene oxide and graphitic carbon nitride nanocomposites assembled by electrostatic attraction forces: Synthesis and characterization. Mater. Chem. Phys. 2019, 228, 228–236. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Åberg, C.; Salvati, A.; Dawson, K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walkey, C.D.; Chan, W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef] [PubMed]
- Doorley, G.W.; Payne, C.K. Cellular binding of nanoparticles in the presence of serum proteins. Chem. Commun. 2011, 47, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Glancy, D.; Zhang, Y.; Wu, J.L.; Ouyang, B.; Ohta, S.; Chan, W.C. Characterizing the protein corona of sub-10 nm nanoparticles. J. Control. Release 2019, 304, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Longin, C.; Petitgonnet, C.; Guilloux-Benatier, M.; Rousseaux, S.; Alexandre, H. Application of flow cytometry to wine microorganisms. Food Microbiol. 2017, 62, 221–231. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malina, T.; Poláková, K.; Hirsch, C.; Svoboda, L.; Zbořil, R. Toxicity of Carbon Nanomaterials—Towards Reliable Viability Assessment via New Approach in Flow Cytometry. Int. J. Mol. Sci. 2021, 22, 7750. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147750
Malina T, Poláková K, Hirsch C, Svoboda L, Zbořil R. Toxicity of Carbon Nanomaterials—Towards Reliable Viability Assessment via New Approach in Flow Cytometry. International Journal of Molecular Sciences. 2021; 22(14):7750. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147750
Chicago/Turabian StyleMalina, Tomáš, Kateřina Poláková, Cordula Hirsch, Ladislav Svoboda, and Radek Zbořil. 2021. "Toxicity of Carbon Nanomaterials—Towards Reliable Viability Assessment via New Approach in Flow Cytometry" International Journal of Molecular Sciences 22, no. 14: 7750. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147750