Dexamethasone Inhibits the Pro-Angiogenic Potential of Primary Human Myoblasts
Abstract
:1. Introduction
2. Results
2.1. Dex Influences Cell Viability Depending on the Presence of VEGF
2.2. Dex Inhibits TLS Formation Depending on VEGF Presence
2.3. The Influence of Dex on Gene Expression Is Independent of the Cells Growing in a 2D or 3D Matrix
2.4. Cell Viability Depends on Exogenous Growth Factors
2.5. Gene Expression Is Affected by Dex and Depends on the Media
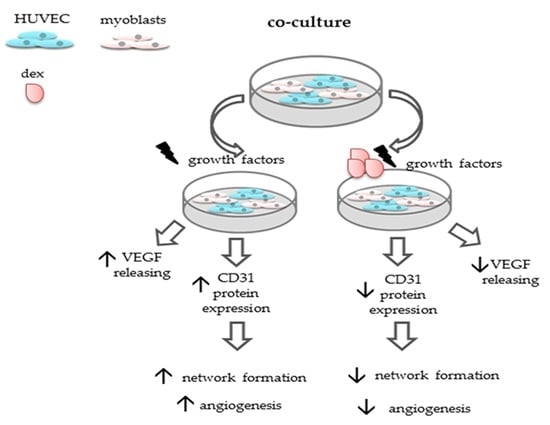
2.6. Designing a Co-Culture System of Human Endothelial Cells with Primary Myoblasts
2.7. Formation of Vessel-Like Structures in the Co-Culture
2.8. Dex Reduces the Angiogenic Potiential of HUVECs in the Co-Culture
2.9. Dex Impairs Angiogenesis and Decreases CD31 Protein Expression in the Co-Culture
2.10. Dex Reduces VEGF Secretion of Myoblast and Decreases Angiogeneses
2.11. Dex Enhances the Expression of Myogenic Transcription Factors but Represses Angiogenesis in the Co-Culture
3. Discussion
3.1. Dex Influence on Endothelial Cells Cultured in Mono-Culture Depends on Different Factors
3.2. Effects of Dex in a Co-Culture System of Human Endothelial Cells with Primary Myoblasts
3.2.1. Formation of Vessel-Like Structures in the Co-Culture
3.2.2. Dex Impairs Angiogenesis
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Cell Viability and Proliferation Assay
4.4. Tube-Like Structure (TLS) Assay
4.5. Evaluation of a Co-Culture System of Primary Human Myoblasts and Endothelial Cells
4.6. Lentiviral Transduction
4.7. Immunofluorescence
4.8. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
4.9. Enzyme-Linked Immunosorbent Assay (ELISA)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liekens, S.; De Clercq, E.; Neyts, J. Angiogenesis: Regulators and Clinical Applications. Biochem. Pharmacol. 2001, 61, 253–270. [Google Scholar] [CrossRef]
- Kerbel, R.S. Tumor Angiogenesis. N. Engl. J. Med. 2008, 358, 2039–2049. [Google Scholar] [CrossRef] [Green Version]
- Chung, A.S.; Ferrara, N. Developmental and Pathological Angiogenesis. Annu. Rev. Cell Dev. Biol. 2011, 27, 563–584. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Molecular Mechanisms and Clinical Applications of Angiogenesis. Nat. Cell Biol. 2011, 473, 298–307. [Google Scholar] [CrossRef] [Green Version]
- Potente, M.; Gerhardt, H.; Carmeliet, P. Basic and Therapeutic Aspects of Angiogenesis. Cell 2011, 146, 873–887. [Google Scholar] [CrossRef] [Green Version]
- Coultas, L.; Chawengsaksophak, K.; Rossant, J. Endothelial Cells and VEGF in Vascular Development. Nat. Cell Biol. 2005, 438, 937–945. [Google Scholar] [CrossRef]
- Ortega, S.; Ittmann, M.; Tsang, S.H.; Ehrlich, M.; Basilico, C. Neuronal Defects and Delayed Wound Healing in Mice Lacking Fibroblast Growth Factor 2. Proc. Natl. Acad. Sci. USA 1998, 95, 5672–5677. [Google Scholar] [CrossRef] [Green Version]
- Nissen, N.N.; Polverini, P.J.; Koch, A.E.; Volin, M.V.; Gamelli, R.L.; DiPietro, L.A. Vascular Endothelial Growth Factor Mediates Angiogenic Activity During the Proliferative Phase of Wound Healing. Am. J. Pathol. 1998, 152, 1445–1452. [Google Scholar] [PubMed]
- Pober, J.S.; Sessa, W. Inflammation and the Blood Microvascular System. Cold Spring Harb. Perspect. Biol. 2015, 7, a016345. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, N.; Sukhova, G.K.; Libby, P.; Reynolds, R.S.; Young, J.L.; Schönbeck, U. Expression of Interleukin (IL)-18 and Functional IL-18 Receptor on Human Vascular Endothelial Cells, Smooth Muscle Cells, and Macrophages. J. Exp. Med. 2002, 195, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Knipe, L.; Meli, A.; Hewlett, L.; Bierings, R.; Dempster, J.; Skehel, P.; Hannah, M.J.; Carter, T. A Revised Model for the Secretion of TPA and Cytokines from Cultured Endothelial Cells. Blood 2010, 116, 2183–2191. [Google Scholar] [CrossRef] [Green Version]
- Mai, J.; Nanayakkara, G.; Lopez-Pastrana, J.; Li, X.; Li, Y.-F.; Wang, X.; Song, A.; Virtue, A.; Shao, Y.; Shan, H.; et al. Interleukin-17A Promotes Aortic Endothelial Cell Activation via Transcriptionally and Post-Translationally Activating p38 Mitogen-Activated Protein Kinase (MAPK) Pathway. J. Biol. Chem. 2016, 291, 4939–4954. [Google Scholar] [CrossRef] [Green Version]
- Chung, A.S.; Lee, J.; Ferrara, N. Targeting the Tumour Vasculature: Insights from Physiological Angiogenesis. Nat. Rev. Cancer 2010, 10, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Claesson-Welsh, L. VEGF Receptor Signal Transduction. Sci. STKE 2001, 112, re21. [Google Scholar] [CrossRef]
- Logie, J.J.; Ali, S.; Marshall, K.M.; Heck, M.M.S.; Walker, B.R.; Hadoke, P.W.F. Glucocorticoid-Mediated Inhibition of Angiogenic Changes in Human Endothelial Cells Is Not Caused by Reductions in Cell Proliferation or Migration. PLoS ONE 2010, 5, e14476. [Google Scholar] [CrossRef] [Green Version]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; Van Oosterom, A.T.; De Bruijn, E.A. Vascular Endothelial Growth Factor and Angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.-K.; Dimberg, A.; Kreuger, J.; Claesson-Welsh, L. VEGF Receptor Signalling? In Control of Vascular Function. Nat. Rev. Mol. Cell Biol. 2006, 7, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Germani, A.; Di Carlo, A.; Mangoni, A.; Straino, S.; Giacinti, C.; Turrini, P.; Biglioli, P.; Capogrossi, M.C. Vascular Endothelial Growth Factor Modulates Skeletal Myoblast Function. Am. J. Pathol. 2003, 163, 1417–1428. [Google Scholar] [CrossRef] [Green Version]
- Joos, G.F.; Brusselle, G.; Van Hoecke, H.; Van Cauwenberge, P.; Bousquet, J.; Pauwels, R.A. Positioning of Glucocorticosteroids in Asthma and Allergic Rhinitis Guidelines (Versus Other Therapies). Immunol. Allergy Clin. N. Am. 2005, 25, 597–612. [Google Scholar] [CrossRef]
- Schäcke, H.; Döcke, W.-D.; Asadullah, K. Mechanisms Involved in the Side Effects of Glucocorticoids. Pharmacol. Ther. 2002, 96, 23–43. [Google Scholar] [CrossRef]
- Shi, J.-X.; Li, J.-S.; Hu, R.; Shi, Y.; Su, X.; Guo, X.-J.; Li, X.-M. Tristetraprolin Is Involved in the Glucocorticoid-Mediated Interleukin 8 Repression. Int. Immunopharmacol. 2014, 22, 480–485. [Google Scholar] [CrossRef] [PubMed]
- You, Q.-H.; Zhang, D.; Niu, C.-C.; Zhu, Z.-M.; Wang, N.; Yue, Y.; Sun, G.-Y. Expression of IL-17A and IL-17F in Lipopolysaccharide-Induced Acute Lung Injury and the Counteraction of Anisodamine or Methylprednisolone. Cytokine 2014, 66, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Zakkar, M.; Le, A.L.; Chaudhury, H.; Ruud, O.; Punjabi, P.P.; Anderson, J.R.; Mullholand, J.W.; Clements, A.T.; Krams, R.; Foin, N.; et al. Dexamethasone Arterializes Venous Endothelial Cells by Inducing Mitogen-Activated Protein Kinase Phosphatase-1: A Novel Antiinflammatory Treatment for Vein grafts? Circulation 2011, 123, 524–532. [Google Scholar] [CrossRef] [Green Version]
- Gelati, M.; Corsini, E.; De Rossi, M.; Masini, L.; Bernardi, G.; Massa, G.; Boiardi, A.; Salmaggi, A. Methylprednisolone Acts on Peripheral Blood Mononuclear Cells and Endothelium in Inhibiting Migration Phenomena in Patients With Multiple Sclerosis. Arch. Neurol. 2002, 59, 774–780. [Google Scholar] [CrossRef]
- Vandevyver, S.; Dejager, L.; Tuckermann, J.; Libert, C. New Insights into the Anti-Inflammatory Mechanisms of Glucocorticoids: An Emerging Role for Glucocorticoid-Receptor-Mediated Transactivation. Endocrinology 2013, 154, 993–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoncini, T.; Maffei, S.; Basta, G.; Barsacchi, G.; Genazzani, A.R.; Liao, J.K.; De Caterina, R. Estrogens and Glucocorticoids Inhibit Endothelial Vascular Cell Adhesion Molecule-1 Expression by Different Transcriptional Mechanisms. Circ. Res. 2000, 87, 19–25. [Google Scholar] [CrossRef]
- Yano, A.; Fujii, Y.; Iwai, A.; Kageyama, Y.; Kihara, K. Glucocorticoids Suppress Tumor Angiogenesis and In Vivo Growth of Prostate Cancer Cells. Clin. Cancer Res. 2006, 12, 3003–3009. [Google Scholar] [CrossRef] [Green Version]
- Gille, J.; Reisinger, K.; Westphal-Varghese, B.; Kaufmann, R. Decreased MRNA Stability As a Mechanism of Glucocorticoid-Mediated Inhibition of Vascular Endothelial Growth Factor Gene Expression by Cultured Keratinocytes. J. Investig. Dermatol. 2001, 117, 1581–1587. [Google Scholar] [CrossRef] [Green Version]
- Pufe, T.; Scholz-Ahrens, K.E.; Franke, A.T.M.; Petersen, W.; Mentlein, R.; Varoga, D.; Tillmann, B.; Schrezenmeir, J.; Glüer, C.C. The Role of Vascular Endothelial Growth Factor in Glucocorticoid-Induced Bone Loss: Evaluation in a Minipig Model. Bone 2003, 33, 869–876. [Google Scholar] [CrossRef]
- Hasselgren, P.-O. Glucocorticoids and Muscle Catabolism. Curr. Opin. Clin. Nutr. Metab. Care 1999, 2, 201–205. [Google Scholar] [CrossRef]
- Annane, D. What Is the Evidence for Harm of Neuromuscular Blockade and Corticosteroid Use in the Intensive Care Unit? Semin. Respir. Crit. Care Med. 2016, 37, 51–56. [Google Scholar] [CrossRef]
- Hermans, G.; Berghe, G.V.D. Clinical Review: Intensive Care Unit Acquired Weakness. Crit. Care 2015, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Langendorf, E.K.; Rommens, P.M.; Drees, P.; Mattyasovszky, S.G.; Ritz, U. Detecting the Effects of the Glucocorticoid Dexamethasone on Primary Human Skeletal Muscle Cells—Differences to the Murine Cell Line. Int. J. Mol. Sci. 2020, 21, 2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, J.R.; McSheehy, P.M.J.; Robinson, S.P.; Troy, H.; Chung, Y.-L.; Leek, R.D.; Williams, K.J.; Stratford, I.J.; Harris, A.L.; Stubbs, M. Metabolic Changes Detected by in Vivo Magnetic Resonance Studies of HEPA-1 Wild-Type Tumors and Tumors Deficient in Hypoxia-Inducible Factor-1beta (HIF-1beta): Evidence of an Anabolic Role for the HIF-1 Pathway. Cancer Res. 2002, 62, 688–695. [Google Scholar] [PubMed]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and Developmental Control of O2 Homeostasis by Hypoxia-Inducible Factor 1alpha. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, A.; Huck, G.; Stiehl, D.; Jelkmann, W.; Hellwig-Bürgel, T. Dexamethasone Impairs Hypoxia-Inducible Factor-1 Function. Biochem. Biophys. Res. Commun. 2008, 372, 336–340. [Google Scholar] [CrossRef]
- Lim, W.; Park, C.; Shim, M.K.; Lee, Y.H.; Lee, Y.M.; Lee, Y. Glucocorticoids Suppress Hypoxia-Induced COX-2 and Hypoxia Inducible Factor-1alpha Expression through the Induction of Glucocorticoid-Induced Leucine Zipper. Br. J. Pharmacol. 2014, 171, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Kodama, T.; Shimizu, N.; Yoshikawa, N.; Makino, Y.; Ouchida, R.; Okamoto, K.; Hisada, T.; Nakamura, H.; Morimoto, C.; Tanaka, H. Role of the Glucocorticoid Receptor for Regulation of Hypoxia-Dependent Gene Expression. J. Biol. Chem. 2003, 278, 33384–33391. [Google Scholar] [CrossRef] [Green Version]
- De Bosscher, K.; Berghe, W.V.; Haegeman, G. The Interplay Between the Glucocorticoid Receptor and Nuclear Factor-KappaB or Activator Protein-1: Molecular Mechanisms for Gene Repression. Endocr. Rev. 2003, 24, 488–522. [Google Scholar] [CrossRef] [Green Version]
- Han, D.-S.; Yang, W.-S.; Kao, T.-W. Dexamethasone Treatment at the Myoblast Stage Enhanced C2C12 Myocyte Differentiation. Int. J. Med. Sci. 2017, 14, 434–443. [Google Scholar] [CrossRef] [Green Version]
- Young, H.Y.S.S.; Woo, J.J. The Effects of Trapa Japonica Fructus Protecs Dexamethasone Induced Muscle Atrophy in C2C12 Myotubes. Int. J. Food Sci. Nutr. Res. 2019, 2, 9. [Google Scholar]
- Wang, R.; Jiao, H.; Zhao, J.; Wang, X.; Lin, H. Glucocorticoids Enhance Muscle Proteolysis through a Myostatin-Dependent Pathway at the Early Stage. PLoS ONE 2016, 11, e0156225. [Google Scholar] [CrossRef]
- Desler, M.M.; Jones, S.J.; Smith, C.W.; Woods, T.L. Effects of Dexamethasone and Anabolic Agents on Proliferation and Protein Synthesis and Degradation in C2C12 Myogenic Cells. J. Anim. Sci. 1996, 74, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Agon, K.W.; Capel, A.J.; Fleming, J.W.; Player, D.J.; Martin, N.R.W.; Lewis, M.P. Mechanical Loading of Tissue Engineered Skeletal Muscle Prevents Dexamethasone Induced Myotube Atrophy. J. Muscle Res. Cell Motil. 2020, 1–11. [Google Scholar] [CrossRef]
- Shimizu, K.; Genma, R.; Gotou, Y.; Nagasaka, S.; Honda, H. Three-Dimensional Culture Model of Skeletal Muscle Tissue With Atrophy Induced by Dexamethasone. Bioengineering 2017, 4, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J.; Langer, R.; Linhardt, R.; Haudenschild, C.; Taylor, S. Angiogenesis Inhibition and Tumor Regression Caused by Heparin or a Heparin Fragment in the Presence of Cortisone. Science 1983, 221, 719–725. [Google Scholar] [CrossRef]
- Nauck, M.; Karakiulakis, G.; Perruchoud, A.P.; Papakonstantinou, E.; Roth, M. Corticosteroids Inhibit the Expression of the Vascular Endothelial Growth Factor Gene in Human Vascular Smooth Muscle Cells. Eur. J. Pharmacol. 1998, 341, 309–315. [Google Scholar] [CrossRef]
- Wen, F.-Q.; Liu, X.; Manda, W.; Terasaki, Y.; Kobayashi, T.; Abe, S.; Fang, Q.; Ertl, R.; Manouilova, L.; Rennard, S.I. TH2 Cytokine-Enhanced and TGF-Beta-Enhanced Vascular Endothelial Growth Factor Production by Cultured Human Airway Smooth Muscle Cells Is Attenuated by IFN-Gamma and Corticosteroids. J. Allergy Clin. Immunol. 2003, 111, 1307–1318. [Google Scholar] [CrossRef]
- Nagashima, M.; Yoshino, S.; Aono, H.; Takai, M.; Sasano, M. Inhibitory Effects of Anti-Rheumatic Drugs on Vascular Endothelial Growth Factor in Cultured Rheumatoid Synovial Cells. Clin. Exp. Immunol. 1999, 116, 360–365. [Google Scholar] [CrossRef]
- Carolina, E.; Kato, T.; Khanh, V.C.; Moriguchi, K.; Yamashita, T.; Takeuchi, K.; Hamada, H.; Ohneda, O. Glucocorticoid Impaired the Wound Healing Ability of Endothelial Progenitor Cells by Reducing the Expression of CXCR4 in the PGE2 Pathway. Front. Med. 2018, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Mozo, L.; Suarez-Diaz, A.M.; Gutierrez, C. Glucocorticoids up-Regulate Constitutive Interleukin-10 Production by Human Monocytes. Clin. Exp. Allergy 2004, 34, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Malefyt, R.D.W.; Abrams, J.; Bennett, B.; Figdor, C.; De Vries, J.E. Interleukin 10(IL-10) Inhibits Cytokine Synthesis by Human Monocytes: An Autoregulatory Role of IL-10 Produced by Monocytes. J. Exp. Med. 1991, 174, 1209–1220. [Google Scholar] [CrossRef] [Green Version]
- Elsby, L.M.; Donn, R.; Alourfi, Z.; Green, L.M.; Beaulieu, E.; Ray, D.W. Hypoxia and Glucocorticoid Signaling Converge to Regulate Macrophage Migration Inhibitory Factor Gene Expression. Arthritis Rheum. 2009, 60, 2220–2231. [Google Scholar] [CrossRef] [Green Version]
- Von Degenfeld, G.; Banfi, A.; Springer, M.L.; Wagner, R.A.; Jacobi, J.; Ozawa, C.R.; Merchant, M.J.; Cooke, J.; Blau, H.M. Microenvironmental VEGF Distribution Is Critical for Stable and Functional Vessel Growth in Ischemia. FASEB J. 2006, 20, 2657–2659. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, C.R.; Banfi, A.; Glazer, N.; Thurston, G.; Springer, M.L.; Kraft, P.E.; McDonald, D.M.; Blau, H.M. Microenvironmental VEGF Concentration, Not Total Dose, Determines a Threshold Between Normal and Aberrant Angiogenesis. J. Clin. Investig. 2004, 113, 516–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsano, A.; Maidhof, R.; Luo, J.; Fujikara, K.; Konofagou, E.E.; Banfi, A.; Vunjak-Novakovic, G. The Effect of Controlled Expression of VEGF by Transduced Myoblasts in a Cardiac Patch on Vascularization in a Mouse Model of Myocardial Infarction. Biomaterials 2013, 34, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Morita, I.; Onodera, M.; Murota, S.-I. Induction of KDR Expression in Bovine Arterial Endothelial Cells by Thrombin: Involvement of Nitric Oxide. J. Cell. Physiol. 2002, 190, 238–250. [Google Scholar] [CrossRef]
- Nagamori, E.; Ngo, T.X.; Takezawa, Y.; Saito, A.; Sawa, Y.; Shimizu, T.; Okano, T.; Taya, M.; Kino-Oka, M. Network Formation through Active Migration of Human Vascular Endothelial Cells in a Multilayered Skeletal Myoblast Sheet. Biomaterials 2013, 34, 662–668. [Google Scholar] [CrossRef]
- Menconi, M.; Gonnella, P.; Petkova, V.; Lecker, S.; Hasselgren, P.-O. Dexamethasone and Corticosterone Induce Similar, But Not Identical, Muscle Wasting Responses in Cultured L6 and C2C12 Myotubes. J. Cell. Biochem. 2008, 105, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Vujčić, M.T.; Velickovic, N.; Ruzdijic, S. Dexamethasone Treatment Affects Nuclear Glucocorticoid Receptor and Glucocorticoid Response Element Binding Activity in Liver of Rats (Rattus Norvegicus) During Aging. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 148, 463–469. [Google Scholar] [CrossRef]
- Erdeljan, P.; Andrews, M.H.; Macdonald, J.F.; Matthews, S.G. Glucocorticoids and Serotonin Alter Glucocorticoid Receptor MRNA Levels in Fetal Guinea-Pig Hippocampal Neurons, in Vitro. Reprod. Fertil. Dev. 2005, 17, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latroche, C.; Weiss-Gayet, M.; Muller, L.; Gitiaux, C.; Leblanc, P.; Liot, S.; Ben-Larbi, S.; Abou-Khalil, R.; Verger, N.; Bardot, P.; et al. Coupling Between Myogenesis and Angiogenesis During Skeletal Muscle Regeneration Is Stimulated by Restorative Macrophages. Stem Cell Rep. 2017, 9, 2018–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langendorf, E.K.; Klein, A.; Drees, P.; Rommens, P.M.; Mattyasovszky, S.G.; Ritz, U. Exposure to Radial Extracorporeal Shockwaves Induces Muscle Regeneration After Muscle Injury in a Surgical Rat Model. J. Orthop. Res. 2019, 38, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Langendorf, E.K.; Klein, A.; Rommens, P.M.; Drees, P.; Ritz, U.; Mattyasovszky, S.G. Calf Blood Compound (CFC) and Homeopathic Drug Induce Differentiation of Primary Human Skeletal Muscle Cells. Endoscopy 2019, 40, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Mattyasovszky, S.G.; Langendorf, E.K.; Ritz, U.; Schmitz, C.; Schmidtmann, I.; Nowak, T.E.; Wagner, D.; Hofmann, A.; Rommens, P.M.; Drees, P. Exposure to Radial Extracorporeal Shock Waves Modulates Viability and Gene Expression of Human Skeletal Muscle Cells: A Controlled in Vitro Study. J. Orthop. Surg. Res. 2018, 13, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brendel, C.; Mueller-Kuller, U.; Schultze-Strasser, S.; Stein, S.; Chen-Wichmann, L.; Krattenmacher, A.; Kunkel, H.; Dillmann, A.; Antoniou, M.N.; Grez, M. Physiological Regulation of Transgene Expression by a Lentiviral Vector Containing the A2UCOE Linked to a Myeloid Promoter. Gene Ther. 2011, 19, 1018–1029. [Google Scholar] [CrossRef] [PubMed]











| Cell Type | Medium | Formulation |
|---|---|---|
| HUVEC | growth medium (HM1) | 5% FCS, EBM-2, EGM™-2 BulletKit™ |
| HUVEC | growth medium without VEGF (HM1-V) | 5% FCS, EBM-2, EGM™-2 BulletKit™, except VEGF |
| HUVEC | without supplementation (EBM-2) | EBM-2 |
| HUVEC | without growth factors (GF) (EBM-2 Ø GF) | EBM-2 + 5% FCS, ascorbic acid, heparin, gentamicin/amphotericin B |
| Myoblasts | growth medium (MM1) | DMEM/F-12, 10% FCS, 2.5 ng/mL bFGF |
| Myoblasts | differentiation medium (DM) | DMEM/F-12, 5% HS |
| CoCulture | induction medium for myo-or angiogenesis medium (1:2) HM1/DM | EBM-2 + EGM-2 Bullet Kit DMEM/F-12, 5% HS (1:2) |
| growth medium HM1/MM1 (1:2) | EBM-2 + EGM™-2 BulletKit™, DMEM/F-12, 10% FCS, 2.5 ng/mL bFGF |
| 1:2 medium HM1/DM (1:2) | EBM-2 + EGM™-2 BulletKit™, DMEM/F-12, 5% HS |
| differentiation medium (DM) | DMEM/F-12, 5% HS |
| Primer | Sequence |
|---|---|
| GAPDH Acc.# M33197 | FW: cgaccactttgtcaagctca |
| RV: aggggagattcagtgtggtg | |
| CD31 Acc.# NM_000442 | FW: cattggcgtgttgggaagaa |
| RV: gctcatgtttgcctagctcc | |
| VEGF Acc.# M32977 | FW: agatgagcttcctacagcacaac |
| RV: aggacttataccgggatttcttg | |
| IL-6 Acc.# NM_000600 | FW: cacagacagccactcacctc |
| RV: cctcaaactccaaaagacca | |
| IL-10 Acc.# NM_000572 | FW: cgtggagcaggtgaagaatg |
| RV: atagaaatgggggttgaggt | |
| HiF1α Acc.# NM_001243084 | FW: gaaaacttggcaaccttgga |
| RV: atctccgtccctcaacctct | |
| GR Acc.# AB307716 | FW: caaatcagcctttcctcggg |
| RV: ctggcccttcaaatgttgct | |
| MyoD Acc.# X56677.1 | FW: ggggctaggttcagctttct |
| RV: gctctggcaaagcaactctt | |
| MyoG Acc.# NM_002479.5 | FW: gccagactatccccttcctc |
| RV: gaggccgcgttatgataaaa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langendorf, E.K.; Rommens, P.M.; Drees, P.; Ritz, U. Dexamethasone Inhibits the Pro-Angiogenic Potential of Primary Human Myoblasts. Int. J. Mol. Sci. 2021, 22, 7986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22157986
Langendorf EK, Rommens PM, Drees P, Ritz U. Dexamethasone Inhibits the Pro-Angiogenic Potential of Primary Human Myoblasts. International Journal of Molecular Sciences. 2021; 22(15):7986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22157986
Chicago/Turabian StyleLangendorf, Eva K., Pol M. Rommens, Philipp Drees, and Ulrike Ritz. 2021. "Dexamethasone Inhibits the Pro-Angiogenic Potential of Primary Human Myoblasts" International Journal of Molecular Sciences 22, no. 15: 7986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22157986