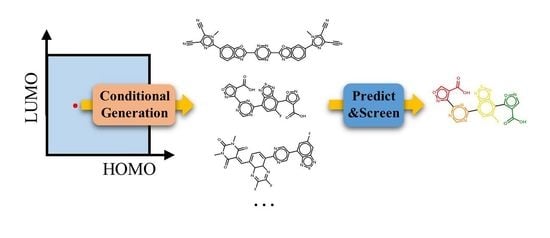
Molecular Conditional Generation and Property Analysis of Non-Fullerene Acceptors with Deep Learning
Abstract
:1. Introduction
2. Methods and Materials
2.1. Molecular Conditional Generative Model
2.2. A Graph-Based Property Prediction Model
2.3. Dataset and Technique Details
3. Results
3.1. Conditional Molecular Generation and Evaluation
3.2. Chemical Space Exploring
3.3. Fragment-Based Molecular Conditional Generation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNN | Convolutional neural network |
| NFA | Non-fullerene acceptor |
| PCE | Power conversion efficiency |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
| SMILES | Simplified molecular input line entry |
| VAE | Variational autoencoder |
| GAN | Generative adversarial network |
| RL | Reinforcement learning |
| TL | Transfer learning |
| RNN | Recurrent neural network |
| GNN | Graph neural network |
| GAT | Graph attention network |
| MMP | Matched molecular pair |
| MAE | Mean absolute error |
Appendix A. The Graph Attention Networks
Appendix B. The Evaluation of CNN-Based Conditional Molecular Generation Models for Benchmarks
| Benchmark | Best of Dataset | SMILES GA | SMILES LSTM | SMILES CNN |
|---|---|---|---|---|
| Celecoxib rediscovery | 0.505 | 0.732 | 1.000 | 0.844 |
| Troglitazone rediscovery | 0.419 | 0.515 | 1.000 | 1.000 |
| Thiothixene rediscovery | 0.456 | 0.598 | 1.000 | 0.687 |
| Aripiprazole similarity | 0.595 | 0.834 | 1.000 | 1.000 |
| Albuterol similarity | 0.719 | 0.907 | 1.000 | 1.000 |
| Mestranol similarity | 0.629 | 0.790 | 1.000 | 0.997 |
| 0.684 | 0.829 | 0.993 | 0.998 | |
| 0.747 | 0.889 | 0.879 | 0.982 | |
| Median molecules 1 | 0.334 | 0.334 | 0.438 | 0.363 |
| Median molecules 2 | 0.351 | 0.380 | 0.422 | 0.377 |
| Osimertinib MPO | 0.839 | 0.886 | 0.907 | 0.863 |
| Fexofenadine MPO | 0.817 | 0.931 | 0.959 | 0.976 |
| Ranolazine MPO | 0.792 | 0.881 | 0.855 | 0.864 |
| Perindopril MPO | 0.575 | 0.661 | 0.808 | 0.679 |
| Amlodipine MPO | 0.696 | 0.722 | 0.894 | 0.763 |
| Sitagliptin MPO | 0.509 | 0.689 | 0.669 | 0.586 |
| Zaleplon MPO | 0.259 | 0.552 | 0.978 | 0.637 |
| Valsartan SMARTS | 0.259 | 0.978 | 0.040 | 0.985 |
| deco hop | 0.933 | 0.970 | 0.996 | 0.983 |
| scaffold hop | 0.738 | 0.885 | 0.998 | 0.844 |
| total | 12.144 | 14.396 | 17.340 | 16.428 |
References
- Servaites, J.D.; Ratner, M.A.; Marks, T.J. Organic Solar Cells: A New Look at Traditional Models. Energy Environ. Sci. 2011, 4, 4410–4422. [Google Scholar] [CrossRef]
- Nielsen, C.B.; Holliday, S.; Chen, H.Y.; Cryer, S.J.; McCulloch, I. Non-Fullerene Electron Acceptors for Use in Organic Solar Cells. Acc. Chem. Res. 2015, 48, 2803–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Barlow, S.; Wang, Z.; Yan, H.; Jen, A.K.Y.; Marder, S.R.; Zhan, X. Non-Fullerene Acceptors for Organic Solar Cells. Nat. Rev. Mater. 2018, 3, 18003. [Google Scholar] [CrossRef]
- Zhang, J.; Tan, H.S.; Guo, X.; Facchetti, A.; Yan, H. Material Insights and Challenges for Non-Fullerene Organic Solar Cells Based on Small Molecular Acceptors. Nat. Energy 2018, 3, 720–731. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, Y.; Jin, K.; Qin, J.; Xu, J.; Li, W.; Xiong, J.; Liu, J.; Xiao, Z.; Sun, K.; et al. 18% Efficiency Organic Solar Cells. Sci. Bull. 2020, 65, 272–275. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Liang, W. Charge Transfer in Organic Molecules for Solar Cells: Theoretical Perspective. Chem. Soc. Rev. 2012, 41, 1075–1087. [Google Scholar] [CrossRef]
- Mater, A.C.; Coote, M.L. Deep Learning in Chemistry. J. Chem. Inf. Model. 2019, 59, 2545–2559. [Google Scholar] [CrossRef]
- Gupta, A.; Müller, A.T.; Huisman, B.J.H.; Fuchs, J.A.; Schneider, P.; Schneider, G. Generative Recurrent Networks for De Novo Drug Design. Mol. Inf. 2018, 37, 1700111. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, T.; Kreisbeck, C.; Becker, J.S.; Aspuru-Guzik, A.; Saikin, S.K. Autonomous Molecular Design: Then and Now. ACS Appl. Mater. Interfaces 2019, 11, 24825–24836. [Google Scholar] [CrossRef]
- Merz, K.M.; De Fabritiis, G.; Wei, G.W. Generative Models for Molecular Design. J. Chem. Inf. Model. 2020, 60, 5635–5636. [Google Scholar] [CrossRef]
- Moosavi, S.M.; Jablonka, K.M.; Smit, B. The Role of Machine Learning in the Understanding and Design of Materials. J. Am. Chem. Soc. 2020, 142, 20273–20287. [Google Scholar] [CrossRef] [PubMed]
- Thiede, L.A.; Krenn, M.; Nigam, A.; Aspuru-Guzik, A. Curiosity in Exploring Chemical Space: Intrinsic Rewards for Deep Molecular Reinforcement Learning. arXiv 2020, arXiv:physics/2012.11293. [Google Scholar]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural Message Passing for Quantum Chemistry. arXiv 2017, arXiv:1704.01212. [Google Scholar]
- Schütt, K.T.; Arbabzadah, F.; Chmiela, S.; Müller, K.R.; Tkatchenko, A. Quantum-Chemical Insights from Deep Tensor Neural Networks. Nat. Commun. 2017, 8, 13890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütt, K.T.; Sauceda, H.E.; Kindermans, P.J.; Tkatchenko, A.; Müller, K.R. SchNet – A Deep Learning Architecture for Molecules and Materials. J. Chem. Phys. 2018, 148, 241722. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ye, W.; Zuo, Y.; Zheng, C.; Ong, S.P. Graph Networks as a Universal Machine Learning Framework for Molecules and Crystals. Chem. Mater. 2019, 31, 3564–3572. [Google Scholar] [CrossRef] [Green Version]
- Unke, O.T.; Meuwly, M. PhysNet: A Neural Network for Predicting Energies, Forces, Dipole Moments and Partial Charges. J. Chem. Theory Comput. 2019, 15, 3678–3693. [Google Scholar] [CrossRef] [Green Version]
- Korolev, V.; Mitrofanov, A.; Korotcov, A.; Tkachenko, V. Graph Convolutional Neural Networks as “General-Purpose” Property Predictors: The Universality and Limits of Applicability. J. Chem. Inf. Model. 2020, 60, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Louis, S.Y.; Zhao, Y.; Nasiri, A.; Wang, X.; Song, Y.; Liu, F.; Hu, J. Graph Convolutional Neural Networks with Global Attention for Improved Materials Property Prediction. Phys. Chem. Chem. Phys. 2020, 22, 18141–18148. [Google Scholar] [CrossRef]
- Brown, N.; McKay, B.; Gilardoni, F.; Gasteiger, J. A Graph-Based Genetic Algorithm and Its Application to the Multiobjective Evolution of Median Molecules. J. Chem. Inf. Comput. Sci. 2004, 44, 1079–1087. [Google Scholar] [CrossRef]
- Ikebata, H.; Hongo, K.; Isomura, T.; Maezono, R.; Yoshida, R. Bayesian Molecular Design with a Chemical Language Model. J. Comput.-Aided Mol. Des. 2017, 31, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Blaschke, T.; Olivecrona, M.; Engkvist, O.; Bajorath, J.; Chen, H. Application of Generative Autoencoder in De Novo Molecular Design. Mol. Inf. 2018, 37, 1700123. [Google Scholar] [CrossRef] [Green Version]
- Jensen, J.H. A Graph-Based Genetic Algorithm and Generative Model/Monte Carlo Tree Search for the Exploration of Chemical Space. Chem. Sci. 2019, 10, 3567–3572. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction Tree Variational Autoencoder for Molecular Graph Generation. arXiv 2019, arXiv:1802.04364. [Google Scholar]
- Kojima, R.; Ishida, S.; Ohta, M.; Iwata, H.; Honma, T.; Okuno, Y. kGCN: A Graph-Based Deep Learning Framework for Chemical Structures. J. Cheminform. 2020, 12, 32. [Google Scholar] [CrossRef]
- Polykovskiy, D.; Zhebrak, A.; Sanchez-Lengeling, B.; Golovanov, S.; Tatanov, O.; Belyaev, S.; Kurbanov, R.; Artamonov, A.; Aladinskiy, V.; Veselov, M.; et al. Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models. arXiv 2020, arXiv:1811.12823. [Google Scholar]
- Weininger, D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Gómez-Bombarelli, R.; Wei, J.N.; Duvenaud, D.; Hernández-Lobato, J.M.; Sánchez-Lengeling, B.; Sheberla, D.; Aguilera-Iparraguirre, J.; Hirzel, T.D.; Adams, R.P.; Aspuru-Guzik, A. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 2018, 4, 268–276. [Google Scholar] [CrossRef]
- Lim, J.; Ryu, S.; Kim, J.W.; Kim, W.Y. Molecular Generative Model Based on Conditional Variational Autoencoder for de Novo Molecular Design. J. Cheminform. 2018, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Cho, K. Conditional Molecular Design with Deep Generative Models. J. Chem. Inf. Model. 2019, 59, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Guimaraes, G.L.; Sanchez-Lengeling, B.; Outeiral, C.; Farias, P.L.C.; Aspuru-Guzik, A. Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models. arXiv 2018, arXiv:1705.10843. [Google Scholar]
- Olivecrona, M.; Blaschke, T.; Engkvist, O.; Chen, H. Molecular De-Novo Design through Deep Reinforcement Learning. J. Cheminform. 2017, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Popova, M.; Isayev, O.; Tropsha, A. Deep Reinforcement Learning for de Novo Drug Design. Sci. Adv. 2018, 4, eaap7885. [Google Scholar] [CrossRef] [Green Version]
- Segler, M.H.S.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Amabilino, S.; Pogány, P.; Pickett, S.D.; Green, D.V.S. Guidelines for Recurrent Neural Network Transfer Learning-Based Molecular Generation of Focused Libraries. J. Chem. Inf. Model. 2020, 60, 5699–5713. [Google Scholar] [CrossRef]
- Yasonik, J. Multiobjective de Novo Drug Design with Recurrent Neural Networks and Nondominated Sorting. J. Cheminform. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotsias, P.C.; Arús-Pous, J.; Chen, H.; Engkvist, O.; Tyrchan, C.; Bjerrum, E.J. Direct Steering of de Novo Molecular Generation with Descriptor Conditional Recurrent Neural Networks. Nat. Mach. Intell. 2020, 2, 254–265. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Merity, S.; Keskar, N.S.; Socher, R. Regularizing and Optimizing LSTM Language Models. arXiv 2017, arXiv:1708.02182. [Google Scholar]
- Bai, S.; Kolter, J.Z.; Koltun, V. An Empirical Evaluation of Generic Convolutional and Recurrent Networks for Sequence Modeling. arXiv 2018, arXiv:1803.01271. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Lopez, S.A.; Sanchez-Lengeling, B.; de Goes Soares, J.; Aspuru-Guzik, A. Design Principles and Top Non-Fullerene Acceptor Candidates for Organic Photovoltaics. Joule 2017, 1, 857–870. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.H. Robust Random Forest Based Non-Fullerene Organic Solar Cells Efficiency Prediction. Org. Electron. 2020, 76, 105465. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Sun, R.; Min, J. Machine Learning for Accelerating the Discovery of High-Performance Donor/Acceptor Pairs in Non-Fullerene Organic Solar Cells. Npj Comput. Mater. 2020, 6, 120. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Omar, Ö.H.; Padula, D.; Geng, Y.; Troisi, A. Computational Identification of Novel Families of Nonfullerene Acceptors by Modification of Known Compounds. J. Phys. Chem. Lett. 2021, 12, 5009–5015. [Google Scholar] [CrossRef]
- Mahmood, A.; Wang, J.L. A Time and Resource Efficient Machine Learning Assisted Design of Non-Fullerene Small Molecule Acceptors for P3HT-Based Organic Solar Cells and Green Solvent Selection. J. Mater. Chem. A 2021, 9, 15684–15695. [Google Scholar] [CrossRef]
- Peng, S.P.; Zhao, Y. Convolutional Neural Networks for the Design and Analysis of Non-Fullerene Acceptors. J. Chem. Inf. Model. 2019, 59, 4993–5001. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.; Fiscato, M.; Segler, M.H.; Vaucher, A.C. GuacaMol: Benchmarking Models for de Novo Molecular Design. J. Chem. Inf. Model. 2019, 59, 1096–1108. [Google Scholar] [CrossRef]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Li, M.; Zhou, J.; Hu, J.; Fan, W.; Zhang, Y.; Gu, Y.; Karypis, G. DGL-LifeSci: An Open-Source Toolkit for Deep Learning on Graphs in Life Science. arXiv 2021, arXiv:q-bio/2106.14232. [Google Scholar]
- RDKit. Available online: https://www.rdkit.org/ (accessed on 4 May 2021).
- Wang, M.; Zheng, D.; Ye, Z.; Gan, Q.; Li, M.; Song, X.; Zhou, J.; Ma, C.; Yu, L.; Gai, Y.; et al. Deep Graph Library: A Graph-Centric, Highly-Performant Package for Graph Neural Networks. arXiv 2020, arXiv:1909.01315. [Google Scholar]
- Dalke, A. Mmpdb: An Open-Source Matched Molecular Pair Platform for Large Multiproperty Data Sets. J. Chem. Inf. Model. 2018, 58, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Leach, A.G.; Jones, H.D.; Cosgrove, D.A.; Kenny, P.W.; Ruston, L.; MacFaul, P.; Wood, J.M.; Colclough, N.; Law, B. Matched Molecular Pairs as a Guide in the Optimization of Pharmaceutical Properties; a Study of Aqueous Solubility, Plasma Protein Binding and Oral Exposure. J. Med. Chem. 2006, 49, 6672–6682. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. A.03. 2016. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| No. | SMILES | Similarity | HOMO Energy (eV) | LUMO Energy (eV) |
|---|---|---|---|---|
| 1 | N#Cc1ccc2c(c1)cc(s2)c1c(F)c(F)c(c2c1nsn2)c1ncncn1 | 0.75 | −6.614 | −3.521 |
| 2 | N#Cc1ccc2c(c1)c(cs2)OC(=O)c1noc(c1C(=O)O)C1=CC(=C(C#N)C#N)C=CO1 | 0.76 | −6.698 | −3.508 |
| 3 | ">N#Cc1ccc2c(ccs2)c1-c1nc(-c2sc(C#N)cc2C#N)ncc1 | 0.78 | −6.541 | −3.260 |
| 4 | N#Cc1c(cc2c(c1)ccs2)C(=O)c1cocc1-c1c2c3c(ccc2)C(=O)N(C)C(=O)c3cc1 | 0.76 | −6.468 | −3.063 |
| 5 | N#Cc1cc(c2c(c1)ccs2)c1ccc2nc(-c3ncnc(-c4oc5ccccc5n4)n3)oc2c1 | 0.81 | −6.463 | −2.881 |
| Mol | Orbital | Energy_calc (eV) | Energy_pred (eV) | Red | Orange | Yellow | Green |
|---|---|---|---|---|---|---|---|
| (a) | HOMO | −6.537 | −6.614 | 0.446 | 0.386 | 0.168 | |
| LUMO | −3.372 | −3.521 | 0.172 | 0.687 | 0.141 | ||
| (b) | HOMO | −6.499 | −6.540 | 0.194 | 0.379 | 0.299 | 0.129 |
| LUMO | −3.240 | −3.359 | 0.310 | 0.081 | 0.498 | 0.111 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, S.-P.; Yang, X.-Y.; Zhao, Y. Molecular Conditional Generation and Property Analysis of Non-Fullerene Acceptors with Deep Learning. Int. J. Mol. Sci. 2021, 22, 9099. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22169099
Peng S-P, Yang X-Y, Zhao Y. Molecular Conditional Generation and Property Analysis of Non-Fullerene Acceptors with Deep Learning. International Journal of Molecular Sciences. 2021; 22(16):9099. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22169099
Chicago/Turabian StylePeng, Shi-Ping, Xin-Yu Yang, and Yi Zhao. 2021. "Molecular Conditional Generation and Property Analysis of Non-Fullerene Acceptors with Deep Learning" International Journal of Molecular Sciences 22, no. 16: 9099. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22169099