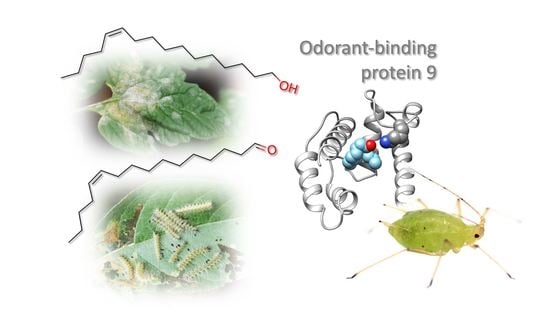
Aphid Odorant-Binding Protein 9 Is Narrowly Tuned to Linear Alcohols and Aldehydes of Sixteen Carbon Atoms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. RNA Extraction, cDNA Synthesis and Cloning
2.3. Preparation of Mutant
2.4. Protein Expression and Purification
2.5. Ligand-Binding Assays
2.6. Modelling and Docking
3. Results
3.1. Expression and Purification
3.2. Ligand-Binding Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Emden, H.F.; Harrington, R. Aphids as Crop Pests, 2nd ed.; CABI Publishing: Wallingford, UK, 2017; ISBN 978-1-78064-709-8. [Google Scholar]
- Miller, G.L.; Foottit, R.G. The Taxonomy of Crop Pests: The Aphids. In Insect Biodiversity; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 627–639. ISBN 978-1-118-94556-8. [Google Scholar]
- Denholm, I.; Pickett, J.A.; Devonshire, A.L.; Devonshire, A.L.; Field, L.M.; Foster, S.P.; Moores, G.D.; Williamson, M.S.; Blackman, R.L. The Evolution of Insecticide Resistance in the Peach–Potato Aphid, Myzus persicae. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1998, 353, 1677–1684. [Google Scholar] [CrossRef] [Green Version]
- Hanson, A.A.; Menger-Anderson, J.; Silverstein, C.; Potter, B.D.; MacRae, I.V.; Hodgson, E.W.; Koch, R.L. Evidence for Soybean Aphid (Hemiptera: Aphididae) Resistance to Pyrethroid Insecticides in the Upper Midwestern United States. J. Econ. Entomol. 2017, 110, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Riddick, E.W. Volatile and Non-Volatile Organic Compounds Stimulate Oviposition by Aphidophagous Predators. Insects 2020, 11, 683. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Pickett, J.A. Manipulation of Parasitoids for Aphid Pest Management: Progress and Prospects. Pest Manag. Sci. 2003, 59, 149–155. [Google Scholar] [CrossRef]
- Brewer, M.J.; Elliott, N.C. Biological Control of Cereal Aphids in North America and Mediating Effects of Host Plant and Habitat Manipulations. Annu. Rev. Entomol. 2004, 49, 219–242. [Google Scholar] [CrossRef] [Green Version]
- Dunn, L.; Lequerica, M.; Reid, C.R.; Latty, T. Dual Ecosystem Services of Syrphid Flies (Diptera: Syrphidae): Pollinators and Biological Control Agents. Pest Manag. Sci. 2020, 76, 1973–1979. [Google Scholar] [CrossRef]
- Francis, F.; Martin, T.; Lognay, G.; Haubruge, E. Role of (E)-Beta-Farnesene in Systematic Aphid Prey Location by Episyrphus Balteatus Larvae (Diptera: Syrphidae). Eur. J. Entomol. 2005, 102, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Bowers, W.S.; Nault, L.R.; Webb, R.E.; Dutky, S.R. Aphid Alarm Pheromone: Isolation, Identification, Synthesis. Science 1972, 177, 1121–1122. [Google Scholar] [CrossRef]
- Edwards, L.J.; Siddall, J.B.; Dunham, L.L.; Uden, P.; Kislow, C.J. Trans-β-Farnesene, Alarm Pheromone of the Green Peach Aphid, Myzus Persicae (Sulzer). Nature 1973, 241, 126–127. [Google Scholar] [CrossRef]
- Dewhirst, S.Y.; Pickett, J.A.; Hardie, J. Chapter Twenty-Two—Aphid Pheromones. In Vitamins & Hormones; Litwack, G., Ed.; Pheromones; Academic Press: Cambridge, MA, USA, 2010; Volume 83, pp. 551–574. [Google Scholar]
- Gibson, R.W.; Pickett, J.A. Wild Potato Repels Aphids by Release of Aphid Alarm Pheromone. Nature 1983, 302, 608–609. [Google Scholar] [CrossRef]
- Yu, X.-D.; Pickett, J.; Ma, Y.-Z.; Bruce, T.; Napier, J.; Jones, H.D.; Xia, L.-Q. Metabolic Engineering of Plant-Derived (E)-β-Farnesene Synthase Genes for a Novel Type of Aphid-Resistant Genetically Modified Crop PlantsF. J. Integr. Plant Biol. 2012, 54, 282–299. [Google Scholar] [CrossRef]
- Beale, M.H.; Birkett, M.A.; Bruce, T.J.A.; Chamberlain, K.; Field, L.M.; Huttly, A.K.; Martin, J.L.; Parker, R.; Phillips, A.L.; Pickett, J.A.; et al. Aphid Alarm Pheromone Produced by Transgenic Plants Affects Aphid and Parasitoid Behavior. Proc. Natl. Acad. Sci. USA 2006, 103, 10509–10513. [Google Scholar] [CrossRef] [Green Version]
- Bruce, T.J.A.; Aradottir, G.I.; Smart, L.E.; Martin, J.L.; Caulfield, J.C.; Doherty, A.; Sparks, C.A.; Woodcock, C.M.; Birkett, M.A.; Napier, J.A.; et al. The First Crop Plant Genetically Engineered to Release an Insect Pheromone for Defence. Sci. Rep. 2015, 5, 11183. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.I.A.G. Genome Sequence of the Pea Aphid Acyrthosiphon Pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar] [CrossRef] [Green Version]
- Smadja, C.; Shi, P.; Butlin, R.K.; Robertson, H.M. Large Gene Family Expansions and Adaptive Evolution for Odorant and Gustatory Receptors in the Pea Aphid, Acyrthosiphon Pisum. Mol. Biol. Evol. 2009, 26, 2073–2086. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zhou, J.-J.; Liu, J.-T.; Huang, G.-Z.; Xu, W.-Y.; Zhang, Q.; Chen, J.-L.; Zhang, Y.-J.; Li, X.-C.; Gu, S.-H. Integrative Transcriptomic and Genomic Analysis of Odorant Binding Proteins and Chemosensory Proteins in Aphids. Insect Mol. Biol. 2019, 28, 1–22. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble Proteins of Chemical Communication: An Overview across Arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond Chemoreception: Diverse Tasks of Soluble Olfactory Proteins in Insects: Soluble Olfactory Proteins in Insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Iovinella, I.; Dani, F.R.; Pelosi, P.; Wang, G. Chemosensory Proteins: A Versatile Binding Family. In Olfactory Concepts of Insect Control—Alternative to insecticides; Picimbon, J.-F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 147–169. ISBN 978-3-030-05164-8. [Google Scholar]
- Dawson, G.W.; Pickett, J.A.; Smiley, D.W.M. The Aphid Sex Pheromone Cyclopentanoids: Synthesis in the Elucidation of Structure and Biosynthetic Pathways. Bioorg. Med. Chem. 1996, 4, 351–361. [Google Scholar] [CrossRef]
- Birkett, M.A.; Pickett, J.A. Aphid Sex Pheromones: From Discovery to Commercial Production. Phytochemistry 2003, 62, 651–656. [Google Scholar] [CrossRef]
- Qiao, H.; Tuccori, E.; He, X.; Gazzano, A.; Field, L.; Zhou, J.-J.; Pelosi, P. Discrimination of Alarm Pheromone (E)-β-Farnesene by Aphid Odorant-Binding Proteins. Insect Biochem. Mol. Biol. 2009, 39, 414–419. [Google Scholar] [CrossRef]
- Sun, Y.F.; De Biasio, F.; Qiao, H.L.; Iovinella, I.; Yang, S.X.; Ling, Y.; Riviello, L.; Battaglia, D.; Falabella, P.; Yang, X.L.; et al. Two Odorant-Binding Proteins Mediate the Behavioural Response of Aphids to the Alarm Pheromone (E)-ß-Farnesene and Structural Analogues. PLoS ONE 2012, 7, e32759. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Wang, B.; Grossi, G.; Falabella, P.; Liu, Y.; Yan, S.; Lu, J.; Xi, J.; Wang, G. Molecular Basis of Alarm Pheromone Detection in Aphids. Curr. Biol. 2017, 27, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.; Fan, J.; Zhang, Y.; Xu, Q.; Han, Z.; Sun, J.; Chen, J. Identification and Expression Analysis of Candidate Odorant-Binding Protein and Chemosensory Protein Genes by Antennal Transcriptome of Sitobion Avenae. PLoS ONE 2016, 11, e0161839. [Google Scholar] [CrossRef]
- Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.-G.; Wang, M.-Q. An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion Avenae. Int. J. Mol. Sci. 2020, 21, 8331. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.-T.; Zhang, Y.-J.; Chen, J.-L.; Li, X.-C.; Liang, P.; Gao, X.-W.; Zhou, J.-J.; Gu, S.-H. Coordinative Mediation of the Response to Alarm Pheromones by Three Odorant Binding Proteins in the Green Peach Aphid Myzus Persicae. Insect Biochem. Mol. Biol. 2021, 130, 103528. [Google Scholar] [CrossRef]
- Ganassi, S.; Grazioso, P.; De Cristofaro, A.; Fiorentini, F.; Sabatini, M.A.; Evidente, A.; Altomare, C. Long Chain Alcohols Produced by Trichoderma Citrinoviride Have Phagodeterrent Activity against the Bird Cherry-Oat Aphid Rhopalosiphum padi. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Aznar-Fernández, T.; Cimmino, A.; Masi, M.; Rubiales, D.; Evidente, A. Antifeedant Activity of Long-Chain Alcohols, and Fungal and Plant Metabolites against Pea Aphid (Acyrthosiphon Pisum) as Potential Biocontrol Strategy. Nat. Prod. Res. 2019, 33, 2471–2479. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and Associated Resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.-C.; Tegoni, M.; Cambillau, C. Structural Basis of the Honey Bee PBP Pheromone and pH-Induced Conformational Change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a Protein-Small Molecule Docking Web Service Based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehouck, Y.; Kwasigroch, J.M.; Gilis, D.; Rooman, M. PoPMuSiC 2.1: A Web Server for the Estimation of Protein Stability Changes upon Mutation and Sequence Optimality. BMC Bioinform. 2011, 12, 151. [Google Scholar] [CrossRef]
- Cali, K.; Persaud, K.C. Modification of an Anopheles gambiae Odorant Binding Protein to Create an Array of Chemical Sensors for Detection of Drugs. Sci. Rep. 2020, 10, 3890. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Onofrio, C.; Knoll, W.; Pelosi, P. Aphid Odorant-Binding Protein 9 Is Narrowly Tuned to Linear Alcohols and Aldehydes of Sixteen Carbon Atoms. Insects 2021, 12, 741. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12080741
D’Onofrio C, Knoll W, Pelosi P. Aphid Odorant-Binding Protein 9 Is Narrowly Tuned to Linear Alcohols and Aldehydes of Sixteen Carbon Atoms. Insects. 2021; 12(8):741. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12080741
Chicago/Turabian StyleD’Onofrio, Chiara, Wolfgang Knoll, and Paolo Pelosi. 2021. "Aphid Odorant-Binding Protein 9 Is Narrowly Tuned to Linear Alcohols and Aldehydes of Sixteen Carbon Atoms" Insects 12, no. 8: 741. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12080741