1. Introduction
Exhaustive grid parameter search is a widely-used hyperparameter optimization strategy in the context of machine learning [
1]. Typically, it is used to search through a manually-defined subset of hyperparameters of a learning algorithm. It is a simple tool for optimizing the performance of machine learning algorithms and can explore all regions of the defined search space if no local extrema exist, and the surfaces of the parameter combinations are relatively smooth. However, it involves high computational costs, increasing exponentially with the number of hyperparameters, as one predictive model needs to be constructed for each combination of parameters (and possibly for each fold of a Cross–Validation (CV)). It can therefore be extremely time-consuming (taking multiple days, weeks or even months of computation depending on the infrastructure available) even for learning algorithms with a small number of hyperparameters, which is often the case. Random search is another approach that randomly samples parameters in a defined search space. It can also be very time-consuming when working with a large number of hyperparameters and a large number of sample points in the search space. Random search can be more suited if highly local optimal parameter combinations exist that might be missed with grid search. It is a less reproducible approach though. Fortunately, grid, random and similar parameter search paradigms are typically “embarrassingly parallel” (
https://en.wikipedia.org/wiki/Embarrassingly_parallel, as of 18 February 2016) problems, as the computation required for building the predictive model for an individual parameter setting does not depend on the others [
2].
Distributed computing frameworks can help with saving time by running independent tasks simultaneously on multiple computers [
3], including local hardware resources, as well as cloud computing resources. These frameworks can use Central Processing Units (CPUs), Graphical Processing Units (GPUs) (which have received much attention recently, especially in the field of deep learning) or a combination of both. Various paradigms for distributed computing exist: Message Passing Interface (MPI) (
https://en.wikipedia.org/wiki/Message_Passing_Interface, as of 18 February 2016) and related projects, such as Open Multi–Processing (OpenMP) are geared towards shared memory and efficient multi-threading. They are well-suited for large computational problems requiring frequent communication between threads (either on a single computer or over a network) and are classically targeted at languages, such as C, C++ or Fortran. They offer fast performance, but can increase the complexity of software development and require high-performance networking in order to avoid bottlenecks when working with large amounts of data. Other paradigms for large-scale data processing, including MapReduce implementations, such as Apache Hadoop (
http://hadoop.apache.org/, as of 18 February 2016), are more aimed towards data locality, fault tolerance, commodity hardware and simple programming (with a stronger link to languages, such as Java or Python). They are more suited for the parallelization of general computation or data processing tasks, with specific tools available for different kinds of processing (for example, Apache Spark (
http://spark.apache.org/, as of 18 February 2016) for in-memory processing or Apache Storm (
http://storm.apache.org/, as of 18 February 2016) for real-time stream-based computation). All of these frameworks are commonly used in medical imaging and machine learning research [
3,
4].
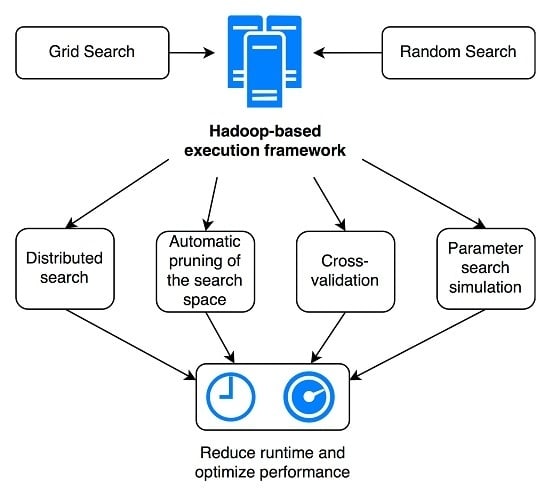
It is also noteworthy to mention that although hyperparameter search should be as exhaustive as possible, there often exist large areas of the search domain that produce suboptimal results, therefore offering opportunities to intelligently reduce the search space and computation time. In a distributed setting, this can complicate the process, as the pruning operation requires sharing information between tasks. To this end, a distributed synchronization mechanism can be designed to allow identifying parameter combinations yielding suboptimal results and subsequently canceling their execution in order to further decrease the total computational time. Moreover, parameter search can be a lengthy process, even when executed within a distributed environment. Therefore, the availability of a parallel execution simulation tool can help estimate the total runtime for varying conditions, such as the number of available computation tasks. Such a simulation tool can also be useful for price estimation when using “pay-as-you-go” computing resources in the cloud (most cloud providers offer specific Hadoop instance types and simple cluster setup tools). This allows making a trade-off between the expected optimization of parameters vs. the related costs.
In this article, we present a novel practical framework for the simulation, optimization and execution of parallel parameter search for machine learning algorithms in the context of medical image analysis. It combines all of the aspects discussed above: (i) parallel execution of parameter search, (ii) intelligent identification and cancellation of suboptimal parameter combinations within the distributed environment and (iii) simulation of the total parallel runtime according to the number of computing nodes available when executed in a distributed architecture. The objective is to allow easily running very fine-grained grid or random parameter search experiments in a reasonable amount of time, while maximizing the likelihood of finding one of the best-performing parameter combinations. We evaluated our framework with two use-cases in the article: lung tissue identification in Computed Tomography (CT) images using (I) Support Vector Machines (SVMs) based on a Radial Basis Function (RBF) kernel and (II) Random Forests (RFs). Results for both grid and random search strategies are provided. The main contributions of the article concern the practical design, implementation and testing of a distributed parameter optimization framework, leveraging software, such as Hadoop and ZooKeeper, in order to enable efficient distributed execution and synchronization, intelligently monitoring the global evolution of the grid search and canceling poorly-performing tasks based on several user-defined criteria, on real data and with a real problem in a scenario potentially similar to many research groups in data science. This has not been done so far, to the best of our knowledge. A second contribution is the developed simulation tool that allows estimating costs and benefits for a large number of scenarios prior to choosing the solution that is optimal for specific constraints. Compared to other publications with a more theoretical focus on hyperparameter optimization algorithms or system design principles, such as [
2,
5,
6,
7,
8,
9], this paper describes a distributed framework that is already implemented and working and has been tested on medical imaging data as an example application field. Only a small number of parameters were optimized in this case, but the same framework also applies to larger parameter spaces.
The rest of the article is structured as follows:
Section 2 discusses existing projects, tools and articles related to the task of hyperparameter optimization.
Section 3 presents the datasets, existing tools and algorithms that were used. The implementation of the developed framework and the experimental results obtained are detailed in
Section 4. The findings and limitations are discussed in
Section 5. Finally, conclusions are drawn and future work is outlined in
Section 6.
2. Related Work
Extensive research has already been conducted in the field of optimizing and improving on the classical grid parameter search model and achieving more efficient hyperparameter optimization in the context of machine learning applications. In 2002, Chapelle
et al. proposed a method for tuning kernel parameters of SVMs using a gradient descent algorithm [
10]. A method for evolutionary tuning of hyperparameters in SVMs using Gaussian kernels was proposed in [
7]. Bergstra
et al. [
2] showed that using random search instead of a pure grid search (in the same setting) can yield equivalent or better results in a fraction of the computation time. Snoek
et al. proposed methods for performing Bayesian optimization of various machine learning algorithms, which supports parallel execution on multiple cores and can reach or surpass human expert-level optimization in various use-cases [
9]. Bergstra
et al. also proposed novel techniques for hyperparameter optimization using a Gaussian process approach in order to train neural networks and Deep Belief Networks (DBNs). They proposed the Tree–structured Parzen Estimator (TPE) approach and discuss the parallelization of their techniques using GPUs [
11]. These papers discuss more the theoretical aspects of optimization, presenting algorithms, but not concrete implementations on a distributed computing architecture.
An extension to the concept of Sequential Model–Based Optimization (SMBO) was proposed in [
6], allowing for general algorithm configuration in a cluster of computers. The paper’s focus is oriented towards the commercial CPLEX (named for the simplex method as implemented in the C programming language) solution and not an open-source solution, such as Hadoop. Auto-WEKA, described in [
8], goes beyond simply optimizing the hyperparameters of a given machine learning method, allowing for an automatic selection of an efficient algorithm among a wide range of classification approaches, including those implemented in the Waikato Environment for Knowledge Analysis (WEKA) machine learning software, but no distributed architecture is discussed in the article. Another noteworthy publication is the work by Luo [
5], who presents the vision and design concepts (but no description of the implementation) of a system aiming to enable very large-scale machine learning on clinical data, using tools, such as Apache Spark and its Machine Learning library (MLlib). The design includes clinical parameter extraction, feature construction and automatic model selection and tuning, with the goal of allowing healthcare researchers with limited computing expertise to easily build predictive models.
Several tools and frameworks have also been released, such as the SUrrogate MOdeling (SUMO) Toolbox [
12], which enables model selection and hyperparameter optimization. It supports grid or cluster computing, but it is geared towards more traditional grid infrastructures, such as the Sun/Oracle Grid Engine, rather than more modern solutions, such as Apache Hadoop, Apache Spark,
etc. Another example is Hyperopt [
13], a Python library for model selection and hyperparameter optimization that supports distributed execution in a cluster using Mongo DataBase (MongoDB) (
http://mongodb.org/, as of 18 February 2016) for inter-process communication, currently for random search and TPE algorithms (
http://jaberg.github.io/hyperopt/, as of 18 February 2016). It does not take advantage of the robust task scheduling and distributed storage features provided by frameworks like Apache Hadoop. In the field of scalable machine learning, Apache Mahout (
http://mahout.apache.org, as of 18 February 2016) allows running several classification algorithms (such as random forests or hidden Markov models), as well as clustering algorithms (k-means clustering, spectral clustering,
etc.) directly on a Hadoop cluster [
4], but it does not address hyperparameter optimization directly and also does not currently provide implementations for certain important classification algorithms, such as SVMs. The MLlib machine learning library (
http://spark.apache.org/mllib/, as of 18 February 2016) provides similar features, using the Apache Spark processing engine instead of Hadoop. Sparks
et al. describe the Training–supported Predictive Analytic Query planner (TuPAQ) system in [
14], an extension of the Machine Learning base (MLbase) (
http://mlbase.org, as of 2 June 2016) platform, which is based on Apache Spark’s MLlib library. TuPAQ allows automatically finding and training predictive models on an Apache Spark cluster. It does not mention a simulation tool that could help with estimating the costs of running experiments of varying complexity in a cloud environment.
Regarding the early termination of unpromising results (pruning the search space of a parameter search) in a distributed setting, [
15] describes a distributed learning method using the multi-armed bandit approach with multiple players. SMBO can also incorporate criteria based on multi-armed bandits [
11]. This is also related to the early termination approaches proposed in this paper that are based on the first experiments and cutoff parameters based on our experiences.
However, articles describing a distributed parameter search setup in detail, including the framework used and an evaluation with real-world clinical data, are scarce. A previous experiment on a much smaller scale was conducted in [
3], where various medical imaging use-cases were analyzed and accelerated using Hadoop. A more naive termination clause was used in a similar SVM optimization problem, where suboptimal tasks were canceled based on a single decision made after processing a fixed number of patients for each parameter combination, based solely on a reference time set by the fastest task reaching the given milestone. The approach taken in this paper is more advanced and flexible, as it cancels tasks during the whole duration of the job, based on an evolving reference value set by all running tasks.
In this article, we describe a very practical approach in detail, based on the Hadoop framework that is easy to set up and manage in a small computing environment, but also easily scalable for larger experiments and supported by many cloud infrastructure providers if the locally available resources become insufficient.
5. Discussion
Three major observations can be deduced from the experimental results: first of all, the speedup achieved by simply distributing a grid parameter search is very substantial, with the total runtime for the search accelerated by a factor of 141× (RF) and 143× (SVM), when compared to an estimation of a serial execution on a single computer (see
Table 2). It also shows that the total runtime decreases almost linearly as the number of nodes (and therefore, available Map tasks) in the Hadoop cluster increases. Second, adding the accuracy and runtime check and canceling suboptimal parameter combinations allows decreasing the runtime even further, by a factor close to or greater than 2× in both use-cases without any significant impact on the maximum achieved accuracy. It also shows that the framework performed well for two different types of classifiers and with a different number of hyperparameters. Third, the results show that several parameter search strategies are supported and work well with the developed framework. The random search experiments ran slightly faster than the grid search using the same number of points and gave equivalent results both with and without task cancellation (see
Table 3). Moreover, reducing the number of random points yielded equivalent results in a fraction of the time needed for the grid search experiments. Repeated experiments also showed that the variability in terms of runtime and achieved performance is minimal. Random search thus provides an interesting option, also in the simulation tool.
The proposed simulation tool was successfully used to estimate job runtimes using a varying number of tasks, with a relative difference of ~10% between the real-world experiment and the simulation for the
standard run using a smaller number of simultaneous tasks (64; see
Table 4). For the
optimized run, the errors were larger, about ~12.5% when simulating with the original amount of Map tasks and ~30.6% when using the smaller number of tasks. Moreover, the simulation provided insights into the effect of varying the cancellation conditions on the maximum achieved classification accuracy and overall job runtime without requiring running a battery of lengthy Hadoop jobs. The latter can be used to reduce costs when using “pay-as-you-go” computing resources in the cloud, which might in the future become the main computation source for many research departments in any case.
Some limitations of this work include the LOPO CV, which could benefit from an added inner Cross–Validation (CV) performed on the training set, in order to reduce the risk of overfitting. Fortunately, this is entirely possible with our framework and is well-suited for parallelizing the task even further. Another limitation concerns the simulation tool, which currently works based only on the results of a real-world experiment. Although it is still interesting to use it on a small-scale experiment and then to extrapolate the data to a more exhaustive experiment, the tool could benefit from a completely simulated mode, where tasks are generated dynamically using an average runtime of tasks input by the user (and adapted with various factors to better represent the variability in runtime of a given experiment and the execution on a distributed framework).
6. Conclusions
The developed framework allows speeding up hyperparameter optimization for medical image classification significantly and easily (both for grid search and random sampling). The distributed nature of the execution environment is leveraged for reducing the search space and gaining further wall time. The simulation tool allows estimating the runtime and results of medical texture analysis experiments under various conditions, as well as extracting information such as a measure of the time-performance trade-off of varying the cancellation margins. These tools can be used in a large variety of tasks that include both image analysis and machine learning aspects. The system using Hadoop is relatively easy to set up, and we expect that many groups can make such optimizations in a much faster way using the results of this article. Indeed, the dramatic reduction in runtime using only a local computing infrastructure can enable the execution of experiments at a scale that may have been dismissed previously, ensuring one to get the best-possible results in the optimization of classification or similar tasks in a very reasonable amount of time. The simulation environment can also help analyze performance and cost trade-offs when optimizing parameters and potentially using cloud environments, allowing one to give cost estimates.
The framework was evaluated with machine learning algorithms with a small number of hyperparameters (i.e., two for SVMs and three for RFs).
In future work, the framework is planned to be tested with other datasets and more classifiers in order to validate its flexibility, potentially also with approaches, such as deep learning, that can use several million hyperparameters and usually rely on GPU computing [
29], often supported by cloud providers, as well. It is also planned to run comparative and larger-scale experiments on a cloud-computing platform instead of using the local Hadoop infrastructure to compare the influence of a mixed environment on runtime, as this can depend much more on the available bandwidth. More advanced task cancellation criteria could also be implemented (e.g., bandit-based method) to allow for more fine-grained control over the tasks to keep. Moreover, adding more sophisticated parameter search strategies to the framework, such as Bayesian optimization or gradient descent, could help improve the system even further, even though it will increase the complexity.