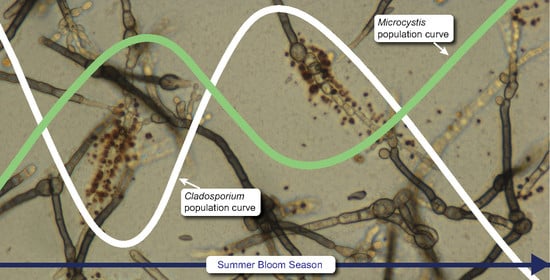
Possible Antagonism between Cladosporium cladosporioides and Microcystis aeruginosa in a Freshwater Lake during Bloom Seasons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Water Sample Collection and Analyses
2.2. Quantification of Microcystis aeruginosa Microcystin-Gene Transcript Copy Number (McyG TCN) in Harsha Lake Water Samples
2.3. Quantification of Cladosporium cladosporioides in Harsha Lake Water Samples
2.4. Cladosporium Spore Counts in Air Samples in the Region of Ohio That Includes Harsha Lake
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carmichael, W.W. Health effects of toxin-producing cyanobacteria: The CyanoHABS. Hum. Ecol. Risk Assess. Int. J. 2001, 7, 1393–1407. [Google Scholar] [CrossRef]
- Leflaive, J.; Ten-Hage, L.O.Ï.C. Algal and cyanobacterial secondary metabolites in freshwaters: A comparison of allelopathic compounds and toxins. Freshw. Biol. 2007, 52, 199–214. [Google Scholar] [CrossRef]
- Pearson, L.; Mihali, T.; Moffitt, M.; Kellmann, R.; Neilan, B. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 2010, 8, 1650–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiegand, C.; Pflugmacher, S. Ecotoxicological effects of selected cyanobacterial secondary metabolites: A short review. Toxicol. Appl. Pharmacol. 2005, 203, 201–218. [Google Scholar] [CrossRef]
- Zurawell, R.W.; Chen, H.; Burke, J.M.; Prepas, E.E. Hepatotoxic cyanobacteria: A review of the biological importance of microcystins in freshwater environments. J. Toxicol. Environ. Health Part B 2005, 8, 1–37. [Google Scholar] [CrossRef]
- Schreidah, C.M.; Ratnayake, K.; Senarath, K.; Karunarathne, A. Microcystins: Biogenesis, toxicity, analysis, and control. Chem. Res. Toxicol. 2020, 33, 2225–2246. [Google Scholar] [CrossRef]
- Omidi, A.; Pflugmacher, S.; Kaplan, A.; Kim, Y.J.; Esterhuizen, M. Reviewing interspecies interactions as a driving force affecting the community structure in lakes via cyanotoxins. Microorganisms 2021, 9, 1583. [Google Scholar] [CrossRef]
- Cooley, D.R.; Mullins, R.F.; Bradley, P.M.; Wilce, R.T. Culture of the upper littoral zone marine alga Pseudendoclonium submarinum induces pathogenic interaction with the fungus Cladosporium cladosporioides. Phycologia 2011, 50, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.D. Fungi as contaminants in indoor air. Atmos. Environ. 1992, 26A, 2163–2172. [Google Scholar] [CrossRef]
- Nix-Stohr, S.; Moshe, R.; Dighton, J. Effects of propagule density and survival strategies on establishment and growth: Further investigations in the phylloplane fungal model system. Micro. Ecol. 2008, 55, 38–44. [Google Scholar] [CrossRef]
- Vesper, S.; Sienkiewicz, N.; Struewing, I.; Linz, D.; Lu, J. Prophylactic addition of glucose suppresses cyanobacterial abundance in lake water. Life 2022, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Allen, J.; Lu, J. Community structures of phytoplankton with emphasis on toxic cyanobacteria in an Ohio inland lake during bloom season. J. Water Resour. Prot. 2017, 9, 1–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Zhu, B.; Struewing, I.; Xu, N.; Duan, S. Nitrogen-phosphorus-associated metabolic activities during the development of a cyanobacterial bloom revealed by metatranscriptomics. Sci. Rep. 2019, 9, 2480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Cao, H.; Li, G.; Du, W.; Xu, G.; Domingo, J.S.; Gu, H.; Xu, N.; Duan, S.; Lu, J. Biodiversity and dynamics of cyanobacterial communities during blooms in temperate lake (Harsha Lake, Ohio, USA). Harmful Algae 2019, 181, 9–18. [Google Scholar] [CrossRef]
- Church, M.J.; Short, C.M.; Jenkins, B.D.; Karl, D.M.; Zehr, J.P. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 2005, 71, 5362–5370. [Google Scholar] [CrossRef] [Green Version]
- Haugland, R.A.; Vesper, S.J. Identification and Quantification of Specific Fungi and Bacteria. U.S. Patent #6,387,652, 14 May 2002. [Google Scholar]
- Haugland, R.A.; Varma, M.; Wymer, L.J.; Vesper, S.J. Quantitative PCR of selected Aspergillus, Penicillium and Paecilomyces species. Syst. Appl. Microbiol. 2004, 27, 198–210. [Google Scholar] [CrossRef]
- Barnard, M.A.; Chaffin, J.D.; Plaas, H.E.; Boyer, G.L.; Wei, B.; Wilhelm, S.W.; Rossignol, K.L.; Braddy, J.S.; Bullerjahn, G.S.; Bridgeman, T.B.; et al. Roles of Nutrient Limitation on Western Lake Erie CyanoHAB Toxin Production. Toxins 2021, 13, 47. [Google Scholar] [CrossRef]
- Wang, K.; Mou, X.; Cao, H.; Struewing, I.; Allen, J.; Lu, J. Co-occurring microorganisms regulate the succession of cyanobacterial harmful algal blooms. Environ. Pollut. 2021, 288, 117682. [Google Scholar] [CrossRef]
- Su, Y.; Hu, M.; Wang, Y.; Zhang, H.; He, C.; Wang, Y.; Wang, D.; Wu, X.; Zhuang, Y.; Hong, S.; et al. Identifying key drivers of harmful algal blooms in a tributary of the Three Gorges Reservoir between different seasons: Causality based on data-driven methods. Environ. Pollut. 2022, 297, 118759. [Google Scholar] [CrossRef]
- Kim, T.; Shin, J.; Lee, D.; Kim, Y.; Na, E.; Park, J.H.; Lim, C.; Cha, Y. Simultaneous feature engineering and interpretation: Forecasting harmful algal blooms using a deep learning approach. Water Res. 2022, 215, 118289. [Google Scholar] [CrossRef]
- Pal, M.; Yesankar, P.J.; Dwivedi, A.; Qureshi, A. Biotic control of harmful algal blooms (HABs): A brief review. J. Environ. Manag. 2020, 268, 10687. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Feng, X.; Jia, Y.; Wang, C.; He, X.; Zhou, Q.; Tian, X. Isolation and evaluation of terrestrial fungi with algicidal ability from Zijin Mountain, Nanjing, China. J. Microbiol. 2011, 49, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Pearson, L.A.; Hisbergues, M.; Börner, T.; Dittmann, E.; Neilan, B.A. Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 2004, 70, 6370–6378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runnegar, M.; Berndt, N.; Kong, S.M.; Lee, E.Y.; Zhang, L. In vivo and in vitro binding of microcystin to protein phosphatases 1 and 2A. Biochem. Biophys. Res. Commun. 1995, 216, 162–169. [Google Scholar] [CrossRef]
- Daub, M.E.; Ehrenshaft, M. The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Ann. Rev. Phyto. 2000, 38, 461–490. [Google Scholar] [CrossRef] [Green Version]
- Schultz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [Green Version]
- Sadaka, N.; Ponge, J.F. Fungal colonization of phyllosphere and litter of Quercus rotundifolia Lam. in a holm oak forest (High Atlas, Morocco). Biol. Fertil. Soils 2003, 39, 30–36. [Google Scholar] [CrossRef]
- O’Donnell, J.; Dickinson, C.H. Pathogenicity of Alternaria and Cladosporium isolates on Phaseolus. Trans. Br. Mycol. Soc. 1980, 74, 335–342. [Google Scholar] [CrossRef]
- Briceno, E.X.; Latorre, B.A. Characterization of Cladosporium rot in grapevines, a problem of growing importance in Chile. Plant Dis. 2008, 92, 1635–1642. [Google Scholar] [CrossRef] [Green Version]


| Study | Sample | Log McyG TCN | SD | Sample | Log CE. C. Clad. Water | SD | Sample | Log No. Clados. Air | SD |
|---|---|---|---|---|---|---|---|---|---|
| Year | Number | Mean | +/− | Number | Mean | +/− | Number | Mean | +/− |
| 2015 | (n = 28) | 2.00 | 0.72 | (n = 28) | 1.90 b | 0.77 | (n = 53) | 2.11 | 0.29 |
| 2016 | (n = 27) | 1.31 a | 0.91 | (n = 27) | 1.33 | 0.54 | (n = 53) | 2.74 c | 0.29 |
| 2017 | (n = 21) | 2.07 | 0.74 | (n = 21) | 1.50 | 0.66 | (n = 53) | 2.14 | 0.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wymer, L.; Vesper, S.; Struewing, I.; Allen, J.; Lu, J. Possible Antagonism between Cladosporium cladosporioides and Microcystis aeruginosa in a Freshwater Lake during Bloom Seasons. Life 2022, 12, 742. https://0-doi-org.brum.beds.ac.uk/10.3390/life12050742
Wymer L, Vesper S, Struewing I, Allen J, Lu J. Possible Antagonism between Cladosporium cladosporioides and Microcystis aeruginosa in a Freshwater Lake during Bloom Seasons. Life. 2022; 12(5):742. https://0-doi-org.brum.beds.ac.uk/10.3390/life12050742
Chicago/Turabian StyleWymer, Larry, Stephen Vesper, Ian Struewing, Joel Allen, and Jingrang Lu. 2022. "Possible Antagonism between Cladosporium cladosporioides and Microcystis aeruginosa in a Freshwater Lake during Bloom Seasons" Life 12, no. 5: 742. https://0-doi-org.brum.beds.ac.uk/10.3390/life12050742