Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. Copper Tolerance and Growth Assessment for the Bacterial Strains
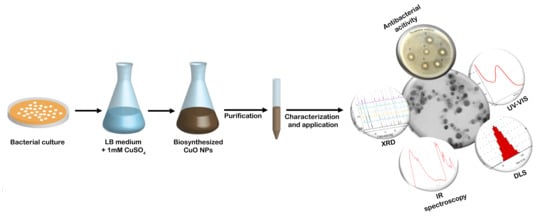
2.2. Biosynthesis of CuNPs
2.3. Ultraviolet–Visible Absorption Spectroscopy (UV–Vis), Dynamic Light Scattering (DLS) Analysis, and Zeta Potential Measurements of CuNPs
2.4. XRD and FTIR Analyses
2.5. Transmission Electron Microscopy (TEM)
2.6. Antimicrobial Activity of the Bacterial CuONPs
3. Materials and Methods
3.1. Culture and Chemicals
3.2. Determination of Copper Maximum Tolerated Concentrations (MTCs)
3.3. Estimation of Bacterial Growth Inhibition by Copper
3.4. Biosynthesis of CuNPs
3.5. Purification of CuNPs
3.6. Dynamic Light Scattering, Zeta Potential Measurement, Transmission Electron Microscopy (TEM), X-ray Diffraction Analysis (XRD), and Fourier-Transform Infrared Spectroscopy (FTIR) Analyses
3.7. Kirby–Bauer Disk Diffusion Susceptibility Test, Minimum Inhibitory Concentration (MIC), and Minimum Bactericidal Concentration (MBC) Evaluation
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Comments on the X-ray Diffraction (XRD) Structural Analysis
References
- Grigore, M.E.; Biscu, E.R.; Holban, A.M.; Gestal, M.C.; Grumezescu, A.M. Methods of synthesis, properties and biomedical applications of CuO Nanoparticles. Pharmaceuticals 2016, 9, 75. [Google Scholar] [CrossRef]
- Katwal, R.; Kaur, H.; Sharma, G.; Naushad, M.; Pathania, D. Electrochemical synthesized copper oxide Nanoparticles for enhanced photocatalytic and antimicrobial activity. J. Ind. Eng. Chem. 2015, 31, 173–184. [Google Scholar] [CrossRef]
- Sastry, M.; Ahmad, A.; Khan, M.I.; Kumar, R. Biosynthesis of metal Nanoparticles using fungi and actinomycetes. Curr. Sci. 2003, 85, 162–170. [Google Scholar]
- Mandal, D.; Bolander, M.E.; Mukhopadhyay, D.; Sarkar, G.; Mukherjee, P. The use of microorganisms for the formation of metal nanoparticles and their application. Appl. Microbiol. Biotechnol. 2006, 69, 485–492. [Google Scholar] [CrossRef]
- Rahmatolahzadeh, R.; Aliabadi, M.; Motevalli, K. Cu and CuO nanostructures: Facile hydrothermal synthesis, characterization and photocatalytic activity using new starting reagents. J. Mater. Sci. Mater. Electron. 2017, 28, 148–156. [Google Scholar] [CrossRef]
- Sagadevan, S.; Pal, K.; Chowdhury, Z.Z. Fabrication of CuO nanoparticles for structural, optical and dielectric analysis using chemical precipitation method. J. Mater. Sci. Mater. Electron. 2017, 28, 12591–12597. [Google Scholar] [CrossRef]
- Zayyoun, N.; Bahmad, L.; Laânab, L.; Jaber, B. The effect of pH on the synthesis of stable Cu2O/CuO Nanoparticles by sol–gel method in a glycolic medium. Appl. Phys. A 2016, 122, 488. [Google Scholar] [CrossRef]
- Shi, G.; Bao, Y.; Chen, B.; Xu, J. Phenol hydroxylation over cubic/monoclinic mixed phase CuO Nanoparticles prepared by chemical vapor deposition. React. Kinet. Mech. Cat. 2017, 122, 289–303. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, K.; Xu, D.; Yang, G.; Huang, H.; Nie, F.; Liu, C.; Yang, S. CuO nanostructures: Synthesis, characterization, growth mechanisms, fundamental properties, and applications. Progress Mater. Sci. 2014, 60, 208–337. [Google Scholar] [CrossRef]
- Waris, A.; Din, M.; Ali, A.; Ali, M.; Afridi, S.; Baset, A.; Khan, A.U. A comprehensive review of green synthesis of copper oxide nanoparticles and their diverse biomedical applications. Inorgan. Chem. Commun. 2021, 123, 1387–7003. [Google Scholar] [CrossRef]
- Wu, F.; Harper, B.J.; Crandon, L.E.; Harper, S.L. Assessment of Cu and CuO nanoparticle ecological responses using laboratory small-scale microcosms Environ. Sci. Nano 2020, 7, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Lontie, R. Copper Proteins and Copper Enzymes; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Kimura, T.; Nishioka, H. Intracellular generation of superoxide by copper sulphate in Escherichia coli. Mutat. Res. 1997, 389, 237–242. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Ramasamy, K.P.; Rajasabapathy, R.; Lips, I.; Mohandass, C.; James, R.A. Genomic features and copper biosorption potential of a new Alcanivorax sp. VBW004 isolated from the shallow hydrothermal vent (Azores, Portugal). Genomics 2020, 112, 3268–3273. [Google Scholar] [CrossRef] [PubMed]
- Pucciarelli, S.; Devaraj, R.R.; Mancini, A.; Ballarini, P.; Castelli, M.; Schrallhammer, M.; Petroni, G.; Miceli, C. Microbial consortium associated with the Antarctic marine ciliate Euplotes focardii: An investigation from genomic sequences. Microb. Ecol. 2015, 70, 484–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pucciarelli, S.; La Terza, A.; Ballarini, P.; Barchetta, S.; Yu, T.; Marziale, F.; Passini, V.; Methé, B.; Detrich, H.W., 3rd; Miceli, C. Molecular cold-adaptation of protein function and gene regulation: The case for comparative genomic analyses in marine ciliated protozoa. Mar. Genomics 2009, 2, 57–66. [Google Scholar] [CrossRef]
- John, M.S.; Nagoth, J.A.; Ramasamy, K.P.; Ballarini, P.; Mozzicafreddo, M.; Mancini, A.; Telatin, A.; Liò, P.; Giuli, G.; Natalello, A.; et al. Horizontal gene transfer and silver nanoparticles production in a new Marinomonas strain isolated from the Antarctic psychrophilic ciliate Euplotes focardii. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- John, M.S.; Nagoth, J.A.; Ramasamy, K.P.; Mancini, A.; Giuli, G.; Natalello, A.; Ballarini, P.; Miceli, C.; Pucciarelli, S. Synthesis of bioactive silver nanoparticles by a pseudomonas strain associated with the Antarctic psychrophilic protozoon Euplotes focardii. Mar. Drugs 2020, 18, 38. [Google Scholar] [CrossRef] [Green Version]
- Orlando, M.; Pucciarelli, S.; Lotti, M. Endolysins from Antarctic Pseudomonas Display lysozyme activity at low temperature. Mar. Drugs. 2020, 18, 579. [Google Scholar] [CrossRef]
- Ramasamy, K.P.; Telatin, A.; Mozzicafreddo, M.; Miceli, C.; Pucciarelli, S. Draft genome sequence of a new pseudomonas sp. Strain, ef1, Associated with the psychrophilic Antarctic ciliate Euplotes focardii. Microbiol. Resour. Announ. 2019, 8, e00867-19. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.L.; Dang, N.L.; Kok, Y.Y.; Yap, K.S.I.; Phang, S.M.; Convey, P. Heavy metal pollution in Antarctica and its potential impacts on algae. Polar Sci. 2019, 20, 75–83. [Google Scholar] [CrossRef]
- Barker, P.F.; Thomas, E. Origin, signature and palaeoclimatic influence of the antarctic circumpolar current. Earth-Sci. Rev. 2004, 66, 143–162. [Google Scholar] [CrossRef]
- Bargagli, R. Environmental contamination in Antarctic ecosystems. Sci. Total Environ. 2008, 400, 212–226. [Google Scholar] [CrossRef]
- De Souza, M.J.; Nair, S.; Loka Bharathi, P.A.; Chandramohan, D. Metal and antibiotic-resistance in psychrotrophic bacteria from Antarctic Marine waters. Ecotoxicology 2006, 15, 379–384. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Casella, P.; Bruni, V.; Michaud, L. Response of bacterial isolates from Antarctic shallow sediments towards heavy metals, antibiotics and polychlorinated biphenyls. Ecotoxicology 2013, 22, 240–250. [Google Scholar] [CrossRef]
- Mancini, A.; Pucciarelli, S. Antibiotic activity of the antioxidant drink effective Microorganism-X (EM-X) extracts against common nosocomial pathogens: An in vitro study. Nat. Prod. Res. 2018, 4, 1–6. [Google Scholar] [CrossRef]
- Ramanathan, R.; Field, M.R.; O’Mullane, A.P.; Smooker, P.M.; Bhargava, S.K.; Bansal, V. Aqueous phase synthesis of copper nanoparticles: A link between heavy metal resistance and nanoparticle synthesis ability in bacterial systems. Nanoscale 2013, 5, 2300–2306. [Google Scholar] [CrossRef] [PubMed]
- Honary, S.; Barabadi, H.; Ghara-El Fathabad, B.; Naguib, F. Green synthesis of copper oxide Nanoparticles using Penicillium aurantiogriseum, Penicillium citrinum and Penicillium waksmanii. Digest. J. Nanomat. Bios. 2012, 7, 999–1005. [Google Scholar]
- Yedurkar, S.M.; Maurya, C.B.; Mahanwar, P.A. A biological approach for the synthesis of copper oxide nanoparticles by Ixora Coccinea leaf extract. J. Mat. Environ. Sci. 2017, 8, 1173–1178. [Google Scholar]
- Dhineshbabu, N.R.; Rajendran, V.; Nithyavathy, N.; Vetumperumal, R. Study of structural and optical properties of cupric oxide nanoparticles. Appl. Nanosci. 2016, 6, 933–939. [Google Scholar] [CrossRef] [Green Version]
- Jovanović, Ž.; Radosavljević, A.; Šiljegović, M.; Bibić, N.; Mišković-Stanković, V.; Kačarević-Popović, Z. Structural and optical characteristics of silver/poly(N-vinyl-2-pyrrolidone) nanosystems synthesized by γ-irradiation. Radiat. Phys. Chem. 2012, 81, 1720–1728. [Google Scholar] [CrossRef]
- Datta Majumdar, T.; Singh, M.; Thapa, M.; Dutta, M.; Mukherjee, A.; Ghosh, C.K. Size-dependent antibacterial activity of copper nanoparticles against Xanthomonas oryzae pv. oryzae–A synthetic and mechanistic approach. Colloid Interface Sci. Commun. 2019, 32, 100190. [Google Scholar] [CrossRef]
- Dadi, R.; Azouani, R.; Traore, M.; Mielcarek, C.; Kanaev, A. Antibacterial activity of ZnO and CuO nanoparticles against gram positive and gram negative strains. Mater. Sci. Eng. C 2019, 104, 109968. [Google Scholar] [CrossRef]
- Ramzan, M.; Obodo, R.M.; Mukhtar, S.; Ilyas, S.Z.; Aziz, F.; Thovhogi, N. Green synthesis of copper oxide nanoparticles using Cedrus deodara aqueous extract for antibacterial activity. Mater. Today Proc. 2021, 36, 576–581. [Google Scholar] [CrossRef]
- Applerot, G.; Lellouche, J.; Lipovsky, A.; Nitzan, Y.; Lubart, R.; Gedanken, A.; Banin, E. Understanding the antibacterial mechanism of CuO nanoparticles: Revealing the route of induced oxidative stress. Small 2012, 8, 3326–3337. [Google Scholar] [CrossRef] [PubMed]
- Shuai, C.; Guo, W.; Gao, C.; Yang, Y.; Wu, P.; Feng, P. An nMgO containing scaffold: Antibacterial activity, degradation properties and cell responses. Int. J. Bioprint. 2018, 4, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI Document EP100-S23; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019.




| CuSO4 Concentration (mM) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organisms | 0.0 | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 | 3.5 | 4 | 4.5 | 5 | 5.5 | 6 |
| Marinomonas ef1 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | ++ | ++ | + | − | − |
| Rhodococcus ef1 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | + | − | − | − |
| Pseudomonas ef1 | +++ | +++ | +++ | +++ | ++ | ++ | + | + | − | − | − | − | − |
| Brevundimonas ef1 | +++ | +++ | +++ | +++ | +++ | ++ | ++ | ++ | ++ | + | + | − | − |
| Bacillus ef1 | +++ | +++ | +++ | +++ | +++ | ++ | ++ | + | + | − | − | − | − |
| Organisms | CuSO4 (mM) |
|---|---|
| Marinomonas ef1 | 5 |
| Rhodococcus ef1 | 4.5 |
| Pseudomonas ef1 | 3.5 |
| Brevundimonas ef1 | 5 |
| Bacillus ef1 | 4 |
| Organisms | Zeta Potential (mV) |
|---|---|
| Marinomonas ef1 | –23.2 |
| Rhodococcus ef1 | –33.8 |
| Pseudomonas ef1 | –33.1 |
| Brevundimonas ef1 | –33.6 |
| Bacillus ef1 | –25.1 |
| MM-ef1 | RH-ef1 | BM-ef1 | PM-ef1 | BC-ef1 | CuSO4 | |
|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | ||||||
| SA | 16 ± 0.2 * | 16 ± 0.3 * | 16 ± 0.1 * | 13 ± 0.4 * | 15 ± 0.2 * | 9 ± 0.3 ° |
| Gram-Negative Bacteria | ||||||
| EC | 15 ± 0.2 * | 16 ± 0.4 * | 17 ± 0.2 * | 16 ± 0.3 * | 16 ± 0.2 * | 10 ± 0.1 ° |
| KP | 16 ± 0.1 * | 15 ± 0.1 * | 17 ± 0.3 * | 16 ± 0.4 * | 16 ± 0.3 * | 10 ± 0.3 ° |
| PM | 16 ± 0.3 * | 17 ± 0.4 * | 16 ± 0.4 * | 15 ± 0.4 * | 15 ± 0.2 * | 11 ± 0.2 ° |
| PrM | 15 ± 0.1 * | 15 ± 0.2 * | 14 ± 0.2 * | 15 ± 0.2 * | 15 ± 0.4 * | 10 ± 0.2 ° |
| CK | 16 ± 0.3 * | 15 ± 0.3 * | 15 ± 0.2 * | 16 ± 0.2 * | 16 ± 0.3 * | 10 ± 0.1 ° |
| AB | 16 ± 0.2 * | 17 ± 0.4 * | 15 ± 0.1 * | 16 ± 0.2 * | 15 ± 0.2 * | 11 ± 0.2 ° |
| SM | 15 ± 0.2 * | 15 ± 0.2 * | 15 ± 0.2 * | 14 ± 01 * | 14 ± 0.2 * | 10 ± 0.3 ° |
| Fungi | ||||||
| CA | 18 ± 0.4 * | 16 ± 0.4 * | 16 ± 0.4 * | 17 ± 0.4 * | 18 ± 0.4 * | 11 ± 0.2 ° |
| CP | 17 ± 0.4 * | 19 ± 0.2 * | 17 ± 0.4 * | 16 ± 0.4 * | 16 ± 0.4 * | 11 ± 0.2 ° |
| MM-ef1 | RH-ef1 | BM-ef1 | PM-ef1 | BC-ef1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| Gram Positive Bacteria | ||||||||||
| SA | 12.5 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 25 ± 0.2 | 25 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.4 | 25 ± 0.2 | 12.5 ± 0.4 | 12.5 ± 0.1 |
| Gram Negative Bacteria | ||||||||||
| EC | 12.5 ± 0.2 | 25 ± 0.3 | 12.5 ± 0.4 | 25 ± 0.3 | 25 ± 0.4 | 25 ± 0.3 | 25 ± 0.3 | 25 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 |
| KP | 12.5 ± 0.1 | 25 ± 0.5 | 6.25 ± 0.3 | 12.5 ± 0.2 | 12.5 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 25 ± 0.4 | 6.25 ± 0.2 | 6.25 ± 0.2 |
| PM-sp | 6.25 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 25 ± 0.3 | 12.5 ± 0.1 | 12.5 ± 0.4 | 12.5 ± 0.3 | 12.5 ± 0.4 |
| PM | 3.12 ± 0.1 | 6.25 ± 0.2 | 6.25 ± 0.2 | 12.5 ± 0.1 | 6.25 ± 0.1 | 12.5 ± 0.2 | 6.25 ± 0.2 | 12.5 ± 0.2 | 6.25 ± 0.1 | 12.5 ± 0.1 |
| CK | 6.25 ± 0.2 | 6.25 ± 0.1 | 12.5 ± 0.3 | 12.5 ± 0.2 | 12.5 ± 0.2 | 25 ± 0.4 | 6.25 ± 0.1 | 12.5 ± 0.4 | 12.5 ± 0.4 | 12.5 ± 0.1 |
| AB | 12.5 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.2 | 12.5 ± 0.4 |
| SM | 6.25 ± 0.1 | 12.5 ± 0.4 | 3.12 ± 0.1 | 12.5 ± 0.2 | 6.25 ± 0.1 | 12.5 ± 0.2 | 6.25 ± 0.1 | 12.5 ± 0.2 | 6.25 ± 0.2 | 12.5 ± 0.2 |
| Fungi | ||||||||||
| CA | 25 ± 0.4 | 25 ± 0.5 | 12.5 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 25 ± 0.4 | 25 ± 0.2 | 25 ± 0.4 | 12.5 ± 0.2 | 25 ± 0.3 |
| CP | 12.5 ± 0.4 | 25 ± 0.3 | 25 ± 0.2 | 25 ± 0.2 | 12.5 ± 0.1 | 25 ± 0.2 | 12.5 ± 0.3 | 25 ± 0.1 | 6.25 ± 0.2 | 12.5 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
John, M.S.; Nagoth, J.A.; Zannotti, M.; Giovannetti, R.; Mancini, A.; Ramasamy, K.P.; Miceli, C.; Pucciarelli, S. Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents. Mar. Drugs 2021, 19, 263. https://0-doi-org.brum.beds.ac.uk/10.3390/md19050263
John MS, Nagoth JA, Zannotti M, Giovannetti R, Mancini A, Ramasamy KP, Miceli C, Pucciarelli S. Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents. Marine Drugs. 2021; 19(5):263. https://0-doi-org.brum.beds.ac.uk/10.3390/md19050263
Chicago/Turabian StyleJohn, Maria Sindhura, Joseph Amruthraj Nagoth, Marco Zannotti, Rita Giovannetti, Alessio Mancini, Kesava Priyan Ramasamy, Cristina Miceli, and Sandra Pucciarelli. 2021. "Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents" Marine Drugs 19, no. 5: 263. https://0-doi-org.brum.beds.ac.uk/10.3390/md19050263