Bioactive Peptides from Algae: Traditional and Novel Generation Strategies, Structure-Function Relationships, and Bioinformatics as Predictive Tools for Bioactivity
Abstract
:1. Introduction
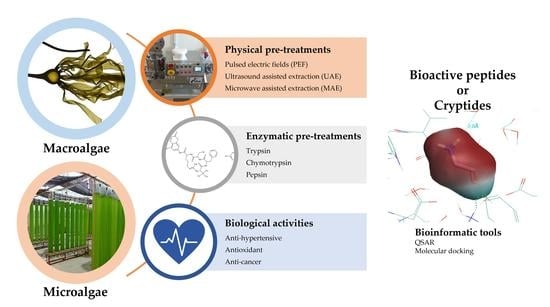
2. Process of Generation and Isolation of Bioactive Peptides
2.1. Pre-Treatments of the Algal Biomass
2.1.1. Physical Pre-Treatments
2.1.2. Enzymatic Pre-Treatments
2.2. Generation of Bioactive Peptides
3. Biological Activities and Modes of Action of Algal Peptides
3.1. Antihypertensive Peptides
3.2. Antioxidant Properties
3.3. Anti-Cancer Properties
4. Application of Novel Bioinformatic Tools
4.1. QSAR
4.2. Molecular Docking
5. Opportunities and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Béné, C.; Barange, M.; Subasinghe, R.; Pinstrup-Andersen, P.; Merino, G.; Hemre, G.-I.; Williams, M. Feeding 9 billion by 2050—Putting fish back on the menu. Food Secur. 2015, 7, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Rösch, C.; Roßmann, M.; Weickert, S. Microalgae for integrated food and fuel production. GCB Bioenergy 2019, 11, 326–334. [Google Scholar] [CrossRef]
- Kusmayadi, A.; Leong, Y.K.; Yen, H.-W.; Huang, C.-Y.; Chang, J.-S. Microalgae as sustainable food and feed sources for animals and humans—Biotechnological and environmental aspects. Chemosphere 2021, 271, 129800. [Google Scholar] [CrossRef]
- Vanthoor-Koopmans, M.; Wijffels, R.H.; Barbosa, M.J.; Eppink, M.H. Biorefinery of microalgae for food and fuel. Bioresour. Technol. 2013, 135, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Cian, R.E.; Drago, S.R.; De Medina, F.S.; Martínez-Augustin, O. Proteins and Carbohydrates from Red Seaweeds: Evidence for Beneficial Effects on Gut Function and Microbiota. Mar. Drugs 2015, 13, 5358–5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safafar, H.; Uldall Nørregaard, P.; Ljubic, A.; Møller, P.; Løvstad Holdt, S.; Jacobsen, C. Enhancement of Protein and Pigment Content in Two Chlorella Species Cultivated on Industrial Process Water. J. Mar. Sci. Eng. 2016, 4, 84. [Google Scholar] [CrossRef] [Green Version]
- Van Krimpen, M.; Bikker, P.; Van der Meer, I.; Van der Peet-Schwering, C.; Vereijken, J. Cultivation, Processing and Nutritional Aspects for Pigs and Poultry of European Protein Sources as Alternatives for Imported Soybean Products; 1570-8616; Wageningen UR Livestock Research: Wageningen, The Netherlands, 2013. [Google Scholar]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Harnedy, P.A.; FitzGerald, R.J. Bioactive proteins, peptides, and amino acids from macroalgae 1. J. Phycol. 2011, 47, 218–232. [Google Scholar] [CrossRef]
- Geada, P.; Moreira, C.; Silva, M.; Nunes, R.; Madureira, L.; Rocha, C.M.R.; Pereira, R.N.; Vicente, A.A.; Teixeira, J.A. Algal proteins: Production strategies and nutritional and functional properties. Bioresour. Technol. 2021, 332, 125125. [Google Scholar] [CrossRef]
- Fleurence, J. The enzymatic degradation of algal cell walls: A useful approach for improving protein accessibility? J. Appl. Phycol. 1999, 11, 313–314. [Google Scholar] [CrossRef]
- Domozych, D.S. Algal Cell Walls. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Fleurence, J.; Massiani, L.; Guyader, O.; Mabeau, S. Use of enzymatic cell wall degradation for improvement of protein extraction from Chondrus crispus, Gracilaria verrucosa and Palmaria palmata. J. Appl. Phycol. 1995, 7, 393. [Google Scholar] [CrossRef]
- Bodénès, P.; Bensalem, S.; Français, O.; Pareau, D.; Le Pioufle, B.; Lopes, F. Inducing reversible or irreversible pores in Chlamydomonas reinhardtii with electroporation: Impact of treatment parameters. Algal Res. 2019, 37, 124–132. [Google Scholar] [CrossRef]
- Buchmann, L.; Brändle, I.; Haberkorn, I.; Hiestand, M.; Mathys, A. Pulsed electric field based cyclic protein extraction of microalgae towards closed-loop biorefinery concepts. Bioresour. Technol. 2019, 291, 121870. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, T.; Rems, L.; Tarek, M.; Miklavčič, D. Membrane electroporation and electropermeabilization: Mechanisms and models. Annu. Rev. Biophys. 2019, 48, 63–91. [Google Scholar] [CrossRef]
- Bensalem, S.; Pareau, D.; Cinquin, B.; Français, O.; Le Pioufle, B.; Lopes, F. Impact of pulsed electric fields and mechanical compressions on the permeability and structure of Chlamydomonas reinhardtii cells. Sci. Rep. 2020, 10, 2668. [Google Scholar] [CrossRef] [Green Version]
- Amorim, M.L.; Soares, J.; Vieira, B.B.; Batista-Silva, W.; Martins, M.A. Extraction of proteins from the microalga Scenedesmus obliquus BR003 followed by lipid extraction of the wet deproteinized biomass using hexane and ethyl acetate. Bioresour. Technol. 2020, 307, 123190. [Google Scholar] [CrossRef]
- Parniakov, O.; Barba, F.J.; Grimi, N.; Marchal, L.; Jubeau, S.; Lebovka, N.; Vorobiev, E. Pulsed electric field and pH assisted selective extraction of intracellular components from microalgae Nannochloropsis. Algal Res. 2015, 8, 128–134. [Google Scholar] [CrossRef]
- Prabhu, M.S.; Levkov, K.; Livney, Y.D.; Israel, A.; Golberg, A. High-voltage pulsed electric field preprocessing enhances extraction of starch, proteins, and ash from marine macroalgae Ulva ohnoi. ACS Sustain. Chem. Eng. 2019, 7, 17453–17463. [Google Scholar] [CrossRef]
- Carullo, D.; Pataro, G.; Donsì, F.; Ferrari, G. Pulsed Electric Fields-Assisted Extraction of Valuable Compounds from Arthrospira Platensis: Effect of Pulse Polarity and Mild Heating. Front. Bioeng. Biotechnol. 2020, 8, 1050. [Google Scholar] [CrossRef]
- Bimakr, M.; Rahman, R.A.; Taip, F.S.; Adzahan, N.M.; Sarker, M.; Islam, Z.; Ganjloo, A. Optimization of ultrasound-assisted extraction of crude oil from winter melon (Benincasa hispida) seed using response surface methodology and evaluation of its antioxidant activity, total phenolic content and fatty acid composition. Molecules 2012, 17, 11748–11762. [Google Scholar] [CrossRef]
- Tiwari, B.K. Ultrasound: A clean, green extraction technology. TrAC Trends Anal. Chem. 2015, 71, 100–109. [Google Scholar] [CrossRef]
- Kadam, S.U.; Álvarez, C.; Tiwari, B.K.; O’Donnell, C.P. Extraction and characterization of protein from Irish brown seaweed Ascophyllum nodosum. Food Res. Int. 2017, 99, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Vernes, L.; Abert-Vian, M.; El Maâtaoui, M.; Tao, Y.; Bornard, I.; Chemat, F. Application of ultrasound for green extraction of proteins from spirulina. Mechanism, optimization, modeling, and industrial prospects. Ultrason. Sonochemistry 2019, 54, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Juin, C.; Chérouvrier, J.-R.; Thiéry, V.; Gagez, A.-L.; Bérard, J.-B.; Joguet, N.; Kaas, R.; Cadoret, J.-P.; Picot, L. Microwave-assisted extraction of phycobiliproteins from Porphyridium purpureum. Appl. Biochem. Biotechnol. 2015, 175, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Grosso, C.; Valentão, P.; Ferreres, F.; Andrade, P.B. Alternative and Efficient Extraction Methods for Marine-Derived Compounds. Mar. Drugs 2015, 13, 3182–3230. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, B.; Christen, P. Recent extraction techniques for natural products: Microwave-assisted extraction and pressurised solvent extraction. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2002, 13, 105–113. [Google Scholar] [CrossRef]
- Pasquet, V.; Chérouvrier, J.-R.; Farhat, F.; Thiéry, V.; Piot, J.-M.; Bérard, J.-B.; Kaas, R.; Serive, B.; Patrice, T.; Cadoret, J.-P.; et al. Study on the microalgal pigments extraction process: Performance of microwave assisted extraction. Process Biochem. 2011, 46, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Al-Zuhair, S.; Ashraf, S.; Hisaindee, S.; Darmaki, N.A.; Battah, S.; Svistunenko, D.; Reeder, B.; Stanway, G.; Chaudhary, A. Enzymatic pre-treatment of microalgae cells for enhanced extraction of proteins. Eng. Life Sci. 2017, 17, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Sierra, L.S.; Dixon, C.K.; Wilken, L.R. Enzymatic cell disruption of the microalgae Chlamydomonas reinhardtii for lipid and protein extraction. Algal Res. 2017, 25, 149–159. [Google Scholar] [CrossRef]
- Paiva, L.; Lima, E.; Neto, A.I.; Baptista, J. Angiotensin I-converting enzyme (ACE) inhibitory activity, antioxidant properties, phenolic content and amino acid profiles of Fucus spiralis L. protein hydrolysate fractions. Mar. Drugs 2017, 15, 311. [Google Scholar] [CrossRef]
- Vásquez, V.; Martínez, R.; Bernal, C. Enzyme-assisted extraction of proteins from the seaweeds Macrocystis pyrifera and Chondracanthus chamissoi: Characterization of the extracts and their bioactive potential. J. Appl. Phycol. 2019, 31, 1999–2010. [Google Scholar] [CrossRef]
- Das, R.S.; Tiwari, B.K.; Garcia-Vaquero, M. Bioactive Peptides from Algae. In Bioactive Peptides from Food: Sources, Analysis, and Functions; CRC Press: Boca Raton, FL, USA, 2022; pp. 31–54. [Google Scholar]
- Siepen, J.A.; Keevil, E.-J.; Knight, D.; Hubbard, S.J. Prediction of missed cleavage sites in tryptic peptides aids protein identification in proteomics. J. Proteome Res. 2007, 6, 399–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, J.; Tymoczko, J.; Stryer, L. Proteases: Facilitating a difficult reaction. Biochemistry 2002. [Google Scholar]
- Brik, A.; Wong, C.-H. HIV-1 protease: Mechanism and drug discovery. Org. Biomol. Chem. 2003, 1, 5–14. [Google Scholar] [CrossRef]
- Available online: https://biosolutions.novozymes.com/en/plant-protein/products/alcalase (accessed on 1 May 2022).
- Tavano, O.L. Protein hydrolysis using proteases: An important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Waglay, A.; Karboune, S. Enzymatic generation of peptides from potato proteins by selected proteases and characterization of their structural properties. Biotechnol. Prog. 2016, 32, 420–429. [Google Scholar] [CrossRef]
- Available online: www.sigmaaldrich.com/IE/en/technical-documents/technical-article/research-and-disease-areas/metabolism-research/papain (accessed on 1 May 2022).
- Available online: http://biocon.es/wp-content/uploads/2016/12/TDS-Bromelain_eng.pdf (accessed on 1 May 2022).
- Available online: https://biosolutions.novozymes.com/en/animal-protein/products/protamex (accessed on 1 May 2022).
- Silva, C.M.; da Fonseca, R.d.S.; Prentice, C. Comparing the hydrolysis degree of industrialization byproducts of Withemouth croaker (Micropogonias furnieri) using microbial enzymes. Int. Food Res. J. 2014, 21, 1757–1761. [Google Scholar]
- Available online: https://www.promega.com/-/media/files/resources/protocols/product-information-sheets/n/elastase-protocol.pdf?rev=996bbf57fd0441709e56af3147154437&sc_lang=en (accessed on 1 May 2022).
- Available online: https://www.promega.com/-/media/files/resources/protocols/product-information-sheets/n/thermolysin-protocol.pdf?rev=4115cf6e496f47afb3be9168515fdc5f&sc_lang=en (accessed on 1 May 2022).
- Gupta, S.S.; Mishra, V.; Mukherjee, M.D.; Saini, P.; Ranjan, K.R. Amino acid derived biopolymers: Recent advances and biomedical applications. Int. J. Biol. Macromol. 2021, 188, 542–567. [Google Scholar] [CrossRef]
- Marcet, I.; Alvarez, C.; Paredes, B.; Diaz, M. Inert and oxidative subcritical water hydrolysis of insoluble egg yolk granular protein, functional properties, and comparison to enzymatic hydrolysis. J. Agric. Food Chem. 2014, 62, 8179–8186. [Google Scholar] [CrossRef]
- Rivas-Vela, C.I.; Amaya-Llano, S.L.; Castaño-Tostado, E.; Castillo-Herrera, G.A. Protein Hydrolysis by Subcritical Water: A New Perspective on Obtaining Bioactive Peptides. Molecules 2021, 26, 6655. [Google Scholar] [CrossRef]
- Fan, X.; Hu, S.; Wang, K.; Yang, R.; Zhang, X. Coupling of ultrasound and subcritical water for peptides production from Spirulina platensis. Food Bioprod. Process. 2020, 121, 105–112. [Google Scholar] [CrossRef]
- Garcia-Moscoso, J.L.; Obeid, W.; Kumar, S.; Hatcher, P.G. Flash hydrolysis of microalgae (Scenedesmus sp.) for protein extraction and production of biofuels intermediates. J. Supercrit. Fluids 2013, 82, 183–190. [Google Scholar] [CrossRef]
- Powell, T.; Bowra, S.; Cooper, H.J. Subcritical water processing of proteins: An alternative to enzymatic digestion? Anal. Chem. 2016, 88, 6425–6432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.-Y.; Zhu, X.; Wan, X.-L.; Fan, Q.; Ma, Y.-H.; Qian, J.; Liu, X.-L.; Shen, Y.-J.; Jiang, J.-H. Hydrolysis technology and kinetics of poultry waste to produce amino acids in subcritical water. J. Anal. Appl. Pyrolysis 2010, 88, 187–191. [Google Scholar] [CrossRef]
- Esteban, M.B.; García, A.J.; Ramos, P.; Márquez, M.C. Sub-critical water hydrolysis of hog hair for amino acid production. Bioresour. Technol. 2010, 101, 2472–2476. [Google Scholar] [CrossRef]
- Melgosa, R.; Marques, M.; Paiva, A.; Bernardo, A.; Fernández, N.; Sá-Nogueira, I.; Simões, P. Subcritical water extraction and hydrolysis of cod (Gadus morhua) frames to produce bioactive protein extracts. Foods 2021, 10, 1222. [Google Scholar] [CrossRef]
- Banerjee, P.; Shanthi, C. Isolation of novel bioactive regions from bovine Achilles tendon collagen having angiotensin I-converting enzyme-inhibitory properties. Process Biochem. 2012, 47, 2335–2346. [Google Scholar] [CrossRef]
- Ahmed, J.; Mulla, M.; Al-Ruwaih, N.; Arfat, Y.A. Effect of high-pressure treatment prior to enzymatic hydrolysis on rheological, thermal, and antioxidant properties of lentil protein isolate. Legume Sci. 2019, 1, e10. [Google Scholar] [CrossRef] [Green Version]
- Adamson, N.J.; Reynolds, E.C. Characterization of casein phosphopeptides prepared using alcalase: Determination of enzyme specificity. Enzym. Microb. Technol. 1996, 19, 202–207. [Google Scholar] [CrossRef]
- Meinlschmidt, P.; Brode, V.; Sevenich, R.; Ueberham, E.; Schweiggert-Weisz, U.; Lehmann, J.; Rauh, C.; Knorr, D.; Eisner, P. High pressure processing assisted enzymatic hydrolysis–An innovative approach for the reduction of soy immunoreactivity. Innov. Food Sci. Emerg. Technol. 2017, 40, 58–67. [Google Scholar] [CrossRef]
- Asaduzzaman, A.K.M.; Chun, B.-S. Recovery of functional materials with thermally stable antioxidative properties in squid muscle hydrolyzates by subcritical water. J. Food Sci. Technol. 2015, 52, 793–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavakoli, O.; Yoshida, H. Conversion of scallop viscera wastes to valuable compounds using sub-critical water. Green Chem. 2006, 8, 100–106. [Google Scholar] [CrossRef]
- Lafarga, T.; Acién-Fernández, F.G.; Garcia-Vaquero, M. Bioactive peptides and carbohydrates from seaweed for food applications: Natural occurrence, isolation, purification, and identification. Algal Res. 2020, 48, 101909. [Google Scholar] [CrossRef]
- Megías, C.; Yust, M.d.M.; Pedroche, J.; Lquari, H.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Purification of an ACE inhibitory peptide after hydrolysis of sunflower (Helianthus annuus L.) protein isolates. J. Agric. Food Chem. 2004, 52, 1928–1932. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In Vitro and In Silico Approaches to Generating and Identifying Angiotensin-Converting Enzyme I Inhibitory Peptides from Green Macroalga Ulva lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daskaya-Dikmen, C.; Yucetepe, A.; Karbancioglu-Guler, F.; Daskaya, H.; Ozcelik, B. Angiotensin-I-Converting Enzyme (ACE)-Inhibitory Peptides from Plants. Nutrients 2017, 9, 316. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and Characterization of Bioactive Pro-Peptides with In Vitro Renin Inhibitory Activities from the Macroalga Palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Aluko, R.E.; Hossain, M.; Rai, D.K.; Hayes, M. Potential of a Renin Inhibitory Peptide from the Red Seaweed Palmaria palmata as a Functional Food Ingredient Following Confirmation and Characterization of a Hypotensive Effect in Spontaneously Hypertensive Rats. J. Agric. Food Chem. 2014, 62, 8352–8356. [Google Scholar] [CrossRef]
- Li, E.C.; Heran, B.S.; Wright, J.M. Angiotensin converting enzyme (ACE) inhibitors versus angiotensin receptor blockers for primary hypertension. Cochrane Database Syst. Rev. 2014, 2014, CD009096. [Google Scholar] [CrossRef]
- Ni, H.; Li, L.; Liu, G.; Hu, S.-Q. Inhibition mechanism and model of an angiotensin I-converting enzyme (ACE)-inhibitory hexapeptide from yeast (Saccharomyces cerevisiae). PLoS ONE 2012, 7, e37077. [Google Scholar] [CrossRef] [Green Version]
- Cooper, W.O.; Hernandez-Diaz, S.; Arbogast, P.G.; Dudley, J.A.; Dyer, S.; Gideon, P.S.; Hall, K.; Ray, W.A. Major congenital malformations after first-trimester exposure to ACE inhibitors. N. Engl. J. Med. 2006, 354, 2443–2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryde, P.G.; Sedman, A.B.; Nugent, C.E.; Barr, M. Angiotensin-converting enzyme inhibitor fetopathy. J. Am. Soc. Nephrol. 1993, 3, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Wijesekara, I. Development and biological activities of marine-derived bioactive peptides: A review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Suetsuna, K.; Chen, J.-R. Identification of antihypertensive peptides from peptic digest of two microalgae, Chlorella vulgaris and Spirulina platensis. Mar. Biotechnol. 2001, 3, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xu, X.; Sun, X.; Zhang, X.; Chen, X.; Xu, N. Preparation and identification of ACE inhibitory peptides from the marine macroalga Ulva intestinalis. Mar. Drugs 2019, 17, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarei, M.; Abidin, N.B.Z.; Auwal, S.M.; Chay, S.Y.; Abdul Haiyee, Z.; Md Sikin, A.; Saari, N. Angiotensin converting enzyme (ACE)-peptide interactions: Inhibition kinetics, in silico molecular docking and stability study of three novel peptides generated from palm kernel cake proteins. Biomolecules 2019, 9, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [Green Version]
- Tadesse, S.A.; Emire, S.A. Production and processing of antioxidant bioactive peptides: A driving force for the functional food market. Heliyon 2020, 6, e04765. [Google Scholar] [CrossRef]
- Canabady-Rochelle, L.L.; Harscoat-Schiavo, C.; Kessler, V.; Aymes, A.; Fournier, F.; Girardet, J.-M. Determination of reducing power and metal chelating ability of antioxidant peptides: Revisited methods. Food Chem. 2015, 183, 129–135. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, D.; Sun, X.; Sun, S.; Xu, N. Preparation and identification of antioxidant peptides from protein hydrolysate of marine alga Gracilariopsis lemaneiformis. J. Appl. Phycol. 2019, 31, 2585–2596. [Google Scholar] [CrossRef]
- Guo, H.; Kouzuma, Y.; Yonekura, M. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009, 113, 238–245. [Google Scholar] [CrossRef]
- Nimalaratne, C.; Bandara, N.; Wu, J. Purification and characterization of antioxidant peptides from enzymatically hydrolyzed chicken egg white. Food Chem. 2015, 188, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-F.; Zhang, Y.Y.; Di He, M.; Li, C.Y.; Zhou, C.X.; Hong, P.Z.; Qian, Z.-J. Antioxidant Peptide Purified from Enzymatic Hydrolysates of Isochrysis Zhanjiangensis and Its Protective Effect against Ethanol Induced Oxidative Stress of HepG2 Cells. Biotechnol. Bioprocess Eng. 2019, 24, 308–317. [Google Scholar] [CrossRef]
- Sheih, I.-C.; Fang, T.J.; Wu, T.-K.; Lin, P.-H. Anticancer and antioxidant activities of the peptide fraction from algae protein waste. J. Agric. Food Chem. 2010, 58, 1202–1207. [Google Scholar] [CrossRef]
- Marqus, S.; Pirogova, E.; Piva, T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017, 24, 21. [Google Scholar] [CrossRef] [Green Version]
- Kakde, D.; Jain, D.; Shrivastava, V.; Kakde, R.; Patil, A. Cancer therapeutics-opportunities, challenges and advances in drug delivery. J. Appl. Pharm. Sci 2011, 1, 1–10. [Google Scholar]
- Murad, H.; Hawat, M.; Ekhtiar, A.; AlJapawe, A.; Abbas, A.; Darwish, H.; Sbenati, O.; Ghannam, A. Induction of G1-phase cell cycle arrest and apoptosis pathway in MDA-MB-231 human breast cancer cells by sulfated polysaccharide extracted from Laurencia papillosa. Cancer Cell Int. 2016, 16, 39. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Bai, L.; Mao, X.; Zhang, X. Novel peptides with anti-proliferation activity from the Porphyra haitanesis hydrolysate. Process Biochem. 2017, 60, 98–107. [Google Scholar] [CrossRef]
- Becerro, M.A.; Goetz, G.; Paul, V.J.; Scheuer, P.J. Chemical Defenses of the Sacoglossan Mollusk Elysia rufescens and Its Host Alga Bryopsis sp. J. Chem. Ecol. 2001, 27, 2287–2299. [Google Scholar] [CrossRef]
- Suárez, Y.; González, L.; Cuadrado, A.; Berciano, M.; Lafarga, M.; Muñoz, A. Kahalalide F, a new marine-derived compound, induces oncosis in human prostate and breast cancer cells. Mol. Cancer Ther. 2003, 2, 863–872. [Google Scholar] [PubMed]
- Wang, B.; Waters, A.L.; Valeriote, F.A.; Hamann, M.T. An efficient and cost-effective approach to kahalalide F N-terminal modifications using a nuisance algal bloom of Bryopsis pennata. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2015, 1850, 1849–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewell, J.; Mayer, I.; Langdon, S.; Smyth, J.; Jodrell, D.; Guichard, S. The mechanism of action of Kahalalide F: Variable cell permeability in human hepatoma cell lines. Eur. J. Cancer 2005, 41, 1637–1644. [Google Scholar] [CrossRef] [PubMed]
- Flores, P.L.; Rodríguez, E.; Zapata, E.; Carbó, R.; Farías, J.M.; Martínez, M. Maitotoxin Is a Potential Selective Activator of the Endogenous Transient Receptor Potential Canonical Type 1 Channel in Xenopus laevis Oocytes. Mar. Drugs 2017, 15, 198. [Google Scholar] [CrossRef] [Green Version]
- Hamann, M.T. Technology evaluation: Kahalalide F, PharmaMar. Curr. Opin. Mol. Ther. 2004, 6, 657. [Google Scholar]
- Available online: https://web.expasy.org/peptide_cutter/ (accessed on 29 March 2022).
- Available online: http://crdd.osdd.net/raghava/toxinpred/ (accessed on 29 March 2022).
- Available online: https://www.ddg-pharmfac.net/AllerTOP/method.html (accessed on 29 March 2022).
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP Database and Other Programs for Processing Bioactive Peptide Sequences. J. AOAC Int. 2019, 91, 965–980. [Google Scholar] [CrossRef] [Green Version]
- Reddy, B.; Jow, T.; Hantash, B.M. Bioactive oligopeptides in dermatology: Part I. Exp. Dermatol. 2012, 21, 563–568. [Google Scholar] [CrossRef]
- Du, Q.S.; Ma, Y.; Xie, N.Z.; Huang, R.B. Two-level QSAR network (2L-QSAR) for peptide inhibitor design based on amino acid properties and sequence positions. SAR QSAR Environ. Res. 2014, 25, 837–851. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Learnings from quantitative structure–activity relationship (QSAR) studies with respect to food protein-derived bioactive peptides: A review. RSC Adv. 2016, 6, 75400–75413. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Singh Chauhan, J.; Nagpal, G.; Kumar, R.; Sharma, M.; Raghava, G.P. An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci. Rep. 2015, 5, 12512. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.U. Descriptors and their selection methods in QSAR analysis: Paradigm for drug design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar]
- Hellberg, S.; Sjoestroem, M.; Skagerberg, B.; Wold, S. Peptide quantitative structure-activity relationships, a multivariate approach. J. Med. Chem. 1987, 30, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.; Eriksson, L.; Jonsson, J.; Sjöström, M.; Wold, S. New Chemical Descriptors Relevant for the Design of Biologically Active Peptides. A Multivariate Characterization of 87 Amino Acids. J. Med. Chem. 1998, 41, 2481–2491. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Singh, H.; Srivastava, H.K.; Singh, S.; Kishore, G.; Raghava, G.P.S. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinform. 2019, 19, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzaei, M.; Mirdamadi, S.; Ehsani, M.R.; Aminlari, M. Production of antioxidant and ACE-inhibitory peptides from Kluyveromyces marxianus protein hydrolysates: Purification and molecular docking. J. Food Drug Anal. 2018, 26, 696–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporgno, M.P.; Mathys, A. Trends in Microalgae Incorporation into Innovative Food Products with Potential Health Benefits. Front. Nutr. 2018, 5, 58. [Google Scholar] [CrossRef] [PubMed]



| Enzyme Name | Type of Enzyme | pH Range | Temperature | Cleavage Preference | References |
|---|---|---|---|---|---|
| Trypsin | Serine protease | 7.8 | 37–42 °C | Positively charged amino acids; R and K | [35] |
| Chymotrypsin | Serine protease | 7.8 | 37–42 °C | Hydrophobic amino acids; Y, F and W | [36] |
| Pepsin | Aspartic protease | 1.25–2.5 | 37–42 °C | Positively charged amino acids; R and K | [37] |
| Alcalase | Serine endopeptidase | 6.5–10 | 60–75 °C | Broad range specificity however, propensity for cleaving aromatic amino acids | [38,39,40] |
| Papain | Cysteine endopeptidase | 6–7 | 65 °C | Broad range specificity, cleaving peptide bonds of basic amino acids, L or G. Papain will not accept V at position 1 and at position 2 prefers large hydrophobic amino acids | [41] |
| Bromaline | Cysteine endopeptidase | 4.5–8 | 35–55 °C | Broad range specificity with preferred cleavage site at the C terminus of K, A, Y and G | [42] |
| Protamex | Mixture of endo- and exo-proteases from Bacillus sp. | 6–9 | 30–65 °C | Broad cleavage range as it is a mixture of proteases | [43,44] |
| Elastase | Serine protease | 9 | 37 °C | Preferred cleavage at the C-terminus of A, V, S, G, L and I | [45] |
| Thermolysin | Metalloproteinase | 5–8.5 | 65–85 °C | Preferred cleavage at the N-terminus of F, V, I, L, M and A | [46] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Connor, J.; Garcia-Vaquero, M.; Meaney, S.; Tiwari, B.K. Bioactive Peptides from Algae: Traditional and Novel Generation Strategies, Structure-Function Relationships, and Bioinformatics as Predictive Tools for Bioactivity. Mar. Drugs 2022, 20, 317. https://0-doi-org.brum.beds.ac.uk/10.3390/md20050317
O’Connor J, Garcia-Vaquero M, Meaney S, Tiwari BK. Bioactive Peptides from Algae: Traditional and Novel Generation Strategies, Structure-Function Relationships, and Bioinformatics as Predictive Tools for Bioactivity. Marine Drugs. 2022; 20(5):317. https://0-doi-org.brum.beds.ac.uk/10.3390/md20050317
Chicago/Turabian StyleO’Connor, Jack, Marco Garcia-Vaquero, Steve Meaney, and Brijesh Kumar Tiwari. 2022. "Bioactive Peptides from Algae: Traditional and Novel Generation Strategies, Structure-Function Relationships, and Bioinformatics as Predictive Tools for Bioactivity" Marine Drugs 20, no. 5: 317. https://0-doi-org.brum.beds.ac.uk/10.3390/md20050317