1. Introduction
The field of membrane biochemistry has seen an exponential expansion in importance, with membrane proteins (MPs) representing 60% of current drug targets [
1]. The native environment for any protein is crucial for revealing physiological mechanisms and functions, especially for membrane proteins, yet this is surprisingly poorly understood. This lack of understanding and use of simplified soluble-phase assays impedes our ability to target membrane proteins using intelligent drug design. Among this functional group of enzymes, oxidoreductases catalyze the oxidation or reduction of quinones in a two-electron, two-proton conversion, generating transmembrane proton (ΔpH) and potential (Δψ) gradients. Electrons are typically funneled from dehydrogenases (e.g., NADH dehydrogenases and succinate dehydrogenases) to terminal oxidases (e.g., cytochrome
bo3;
bd) and finally to terminal electron acceptors (e.g., oxygen) [
2].
When examining membrane-bound oxidoreductase function, the diversity and nature of the lipid environment is a frequently ignored factor. Most biochemical and biophysical studies focus on the use of non-natural lipids in the protein, with phosphatidyl choline c16:0 being the most popular lipid of choice. However, this is rarely the native environment for both the enzymes and the quinones, especially considering that lipid composition can vary dramatically across the three domains of life, and even within a single organism [
3]. For example, the acyl chains of
Escherichia coli lipids change in composition with growth phase and temperature [
4]. It is becoming increasingly obvious that the lipids have a greater role than being mere ‘macroscopic protein supportive structures’ in protein function: cardiolipin (CL) is structurally associated with quinone-utilizing enzymes, such as the cytochrome
bc1 complex [
5] and the type II human dihydroorotate dehydrogenase (DHODH) [
6]. CL is also reported to enhance the catalytic activity of the
Shewanella oneidensis MR-1 MQ-utilizing CymA [
7], the
E. coli UQ-utilizing cytochromes
bo3 and
bd-I, and the
Geobacillus thermodenitrificans (formerly
Bacillus stearothermophilus) and
Corynebacterium glutamicum [
8] cytochrome
bd-I.
Among quinone oxidoreductases, type II NADH dehydrogenases (NDH-2) are considered attractive drug targets due to their absence from higher animal life and their seeming ubiquity among pathogens (e.g.,
Mycobacterium tuberculosis,
E. coli, and
Trypanosoma brucei) [
9]. In contrast with type I NADH dehydrogenases (NDH-1), NDH-2 does not pump protons to create ΔpH, contributing only to membrane electrical potential (Δψ) [
10]. In some organisms, these are the only membrane-bound respiratory enzymes from any electron transport chain known to be responsible for the oxidation of NADH, some even having multiple copies [
10,
11,
12]. Here, we report a study of the model enzyme
Caldalkalibacillus thermarum NDH-2 (
CthNDH-2), which we have previously examined bioelectrochemically using its native quinone, menaquinone-7 (MQ
7) [
10,
11,
12,
13]. Here, we reveal that by changing lipid compositions when assaying a type II NADH:quinone oxidoreductase, phosphatidylethanolamine has an essential role in quinone head group binding and catalysis. We also reveal the importance of acyl chain composition on isoprenoid quinone-mediated catalysis.
2. Materials and Methods
Caldalkalibacillus thermarum NDH-2 was expressed in
E. coli BL21 (DE3) and purified as described previously [
10]. Membranes with overexpressed enzyme at 5 mg/mL (total MP) were solubilized with buffer containing 2% (w/v) n-octyl-β-D-glucopyranoside (OG) (Anatrace, Maumee, OH, USA), among other components. Solubilized membranes were purified by Ni-affinity chromatography afterwards (>96%). Liposomes were prepared as described elsewhere [
10,
14] to a concentration of 10 mg and, where indicated, 1% mass MQ
7. Phosphatidylglycerol (PG; 15:0–18:1), phosphatidylethanolamine (PE; 15:0–18:1), cardiolipin (CL; 16:0–18.1), and
E. coli polar lipid extract (see
Figure 1A,B) suspended in chloroform were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Where indicated, highly hydrophobic MQ
7 (
Figure 1C) was mixed with lipids in chloroform to a final concentration of 1% wt/wt. We note menadione (MQ
0;
Figure 1D) is far less hydrophobic due to the lack of an isoprenoid chemical group and is not integrated into lipids in this manner. These lipids and lipid/quinone mixtures in chloroform were then dried to a film under a nitrogen stream before rehydration in 20 mM MOPS and 30 mM Na
2SO
4 pH 7.4 to a final concentration of 10 mg/mL.
Purified
CthNDH-2 was reconstituted onto lipid bilayer vesicles of various compositions (see
Figure 1B) by autoinsertion–reconstitution [
14] at a concentration of 0.2 mg protein/mL, as described in [
14]. Solution-phase NADH:quinone oxidoreductase activity was performed as described elsewhere [
10,
15], monitoring NADH oxidation by spectroscopy at 340 nm. An extinction coefficient of 6.22 mM
−1cm
−1 was used to calculate NADH concentration, and NDH-2 specific activity was expressed as U.mg protein
−1, where 1 U = 1 μmol NADH oxidized min
−1. Cyclic voltammetry (CV) was carried out as described in [
10]. The preparation of template-stripped gold and the formation of the 8-mercaptooctanol (8MO) self-assembled monolayers (SAMs) were performed as described previously [
10].
3. Results and Discussion
We selected a native
E. coli polar lipid extract (ECPL) due to its well-defined lipid environment, its similarity to the
C. thermarum TA2.A1 lipid head groups [
13], and the commercial availability of its individual lipid components. The
E. coli polar lipid extract (ECPL) consisted of 9.8% cardiolipin (CL), 23.2% phosphatidylglycerol (PG), and 67% phosphatidylethanolamine (PE) (see
Figure 1B). Remarkably, the native lipid extract also had heterogeneous acyl lipid tails, whereas mixtures of synthetic lipids typically have either no variation at all or less variation than the native extract.
Initially, we explored an appropriate solution-phase assay system, which is the ‘gold standard’ of the field for biochemical characterization of quinone-utilizing membrane proteins.
CthNDH-2 was reconstituted in native ECPL and a soluble menaquinone quinone analogue MQ
0 to accept electrons from
CthNDH-2. Since reconstitution occludes the
CthNDH-2 quinone binding site to a degree due to it facing the membrane, we also conducted this assay in the presence and absence of membrane-imbedded menaquinone-7 (1% MQ
7;
Figure 2A,B), the addition of which amounted to picomolar quantities of MQ
7 in this system.
This revealed that not only is MQ
0 capable of entering ECPL to accept electrons from
CthNDH-2, but also the addition of MQ
7 to the vesicles resulted in a ~30% V
max (
Figure 2C). This was not entirely unexpected because, in the system in
Figure 2A, MQ
0 has a barrier to overcome, lowering the diffusion rate past the lipid head groups and into the proteoliposome lipid acyl-chain hydrophobic phase; on the other hand, in the system in
Figure 2B, MQ
0 can readily collect electrons from MQ
7 when the latter is incorporated, which in turn interacts with
CthNDH-2 as it does in cell physiology [
13]. This proposition is supported by a recent molecular dynamics report in which both quinone and quinol molecules showed a strong tendency to localize in the vicinity of the lipid head groups (as shown in
Figure 2B), and translocation of quinones in the bilayer occurred in the 10–100 ns timescale [
16]. While we cannot discount that MQ
0 may also interact with
CthNDH-2 directly in the system in a similar manner to that in
Figure 2A, it is very unlikely because of the diffusion barrier into the proteoliposome, and the sheer speed of quinone in the proteoliposome (i.e., an MQ
0 molecule would more likely ‘meet’ an MQ
7 molecule than a
CthNDH-2 that did not have MQ
7 bound).
The system containing MQ7 is consequently more relevant physiologically and allows us to examine interactions between CthNDH-2 and MQ7. Therefore, we elected to include MQ7 in the proteoliposomes. To further explore the role of lipids in CthNDH-2 kinetics, CthNDH-2 was reconstituted in either native ECPL, 100% PG, 90.2% PG + 9.8% CL, 33% PG + 67% PE, 9.8% CL, 23.2% PG and 67% PE (synthetic ECPL), or native ECPL. As before, we used MQ0 to accept electrons from membrane-imbedded MQ7. This revealed that the catalytic rate for CthNDH-2, and the apparent KM for quinone is heavily influenced by lipid composition.
PG/CL resulted in the slowest catalytic rate (528 μmol·min
−1mg
−1 protein;
Figure 3A,B) and the second highest
KM (53 μM;
Figure 3C). PG resulted in the second slowest NADH catalytic rate (673 μmol·min
−1mg
−1 protein;
Figure 3A,B) and the highest K
M (62.5 μM;
Figure 3C). However, whenever PE is present, K
M is at its lowest and catalytic rates their highest, as observed in the native ECPL lipid environment (
Figure 3A–C). PG + PE resulted in the highest NADH turnover (759 μmol·min
−1mg
−1 protein;
Figure 3A,B), and the lowest K
M values (21.7 μM;
Figure 3C). In the presence of PE, CL had a modest effect on NADH catalytic rate and a severe effect on K
M, while the difference between native ECPL and artificial ECPL was relatively minor (
Figure 3A–C). It would appear that PE is an essential component to achieve maximum
CthNDH-2 catalytic rate, and the
KM 2.5-fold lower than either PG or PG + CL suggests that PE aids in
CthNDH-2 quinone binding.
However, in nature, soluble quinones are not used, so the data presented in
Figure 3 can only tell us about the influence of lipids on quinone head group interactions with
CthNDH-2. Conversely, a native quinone is extremely hydrophobic due to its extensive poly-isoprenoid chain (quinone ‘tail’, see
Figure 1C). To address this, we applied
CthNDH-2, reconstituted in vesicles of various lipid compositions containing 1% MQ
7, to an 8-mercaptooctanol surface attached to a gold electrode (
Figure 4A). Cyclic voltammetry was used to subtract electrons from the MQ
7 pool in the membrane, as NADH was oxidized by
CthNDH-2 (
Figure 4A).
When comparing the results between 100% PG, 90.2% PG + 9.8% CL, 33% PG + 67% PE, 9.8% CL, 23.2% PG, and 67% PE (synthetic ECPL), the trends that were observed using the ‘
Figure 3A,B assay’ approach broadly hold true (
Figure 4B). Yet, in contrast, it is clear that the synthetic ECPL has a far greater
Vmax than a PG/PE mixture, suggesting lipid packing has an influence on
CthNDH-2 quinone catalysis. This is later confirmed when comparing the
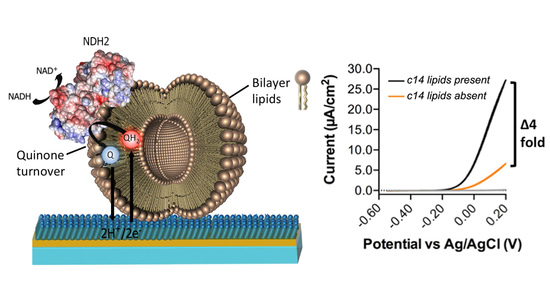
Vmax of synthetic ECPL to native ECPL. Strikingly, the
Vmax of
CthNDH-2 in native ECPL is four-fold higher than in synthetic ECPL (
Figure 4C,D).
In our synthetic ECPL mixture, the acyl chain compositions of the lipids are 15:0–18:1 PG and PE and 16:0–18.1 CL, whereas in native ECPL, this is 14:0–18.1. At face value, this difference in catalytic rate could be attributed to either lipid packing influencing the movement of quinones in membranes or the association with
CthNDH-2. However, since c14 lipids only make up 3.4–3.7% of the acyl chains in native
E. coli polar lipid extracts [
4], the suggestion that diffusion is causative of a four-fold change in activity would seem extremely unlikely. In support of this, the bioelectrochemical hysteresis of the quinone redox peaks is identical between lipid compositions. If a change in MQ
7 diffusion rate was indeed the cause of the four-fold increase in catalytic rate, then the redox peaks would change in potential, whereas here, they do not, as visualized by identical onset potentials (
Figure 4B,C). Clearly, for
CthNDH-2 function with its native substrate MQ
7, acyl chain lengths of 14:0 are of critical interest to explore further. This is not the case for MQ
0.
Collectively, we suggest that the lipid environment enhances the
CthNDH-2 catalytic rate and substrate binding. This indicates that lipids ‘shape’ the binding sites of the quinone head group(s). The notion that lipids can alter
KM is novel; moreover, the influence of PE itself is surprising, considering the history of this CL-interacting protein across domains of life [
5,
6,
7,
8].
Interestingly, both CL and PE are both inverted conical lipids and, in the realms of physical chemistry and lipid biology, are sometimes referred to as ‘non-bilayer lipids’. The reason for this is that in vitro PE promotes the formation of an inverted hexagonal phase instead of a bilayer [
17]. However, most biological membranes contain significant amounts of non-bilayer lipids (e.g.,
E. coli membranes contain a 3:1 ratio of PE:PG, the latter being cylindrical) while being planar bilayers in vivo. In addition, Sendecki et al. [
18] also recently reported the formation of supported lipid bilayers containing up to 90% PE at 37 °C, which is the same temperature as that of the results we report here.
From an observational point of view, the formation of an inverted hexagonal (H
II) phase in vivo has not yet been reported in the literature using modern imaging techniques, such as cryo-electron microscopy, nor has it been unambiguously proven in biological membranes under physiological conditions [
16], although such a phase is thermodynamically favored. However, it should be noted that transient, local, non-bilayer structures have been reported in some species when stressed (e.g., in
E. coli [
4] or
Acholeplasma laidlawii [
19]), where their membranes maintain a beneficial state of quasi-bilayer/non-bilayer phase transition.
Lastly, we also reveal for the first time the profound influence of acyl chain composition on
CthNDH-2 function, a finding we are not aware of being presented before in oxidoreductase biology. Since the subtle chain length difference has such a profound effect, we suggest it might be related to MQ
7 binding and mode of action around the enzyme. This would also strengthen the notion that isoprenoid chain lengths of quinones play an important role in substrate arrangement, as demonstrated for the
S. cerevisiae NDH-2, where either a single (UQ
2, 2 isoprenoid units) or two quinones (UQ
4, 4 isoprenoid units) were bound to structures of the same protein [
11,
12]. Interestingly, gel-phase microdomains in lipid membranes (which are heavily influenced by acyl chain saturation and length) can have dramatic effects on the redox properties of ubiquinone-10 [
20]. These matters collectively require further exploration to discern highly specific concrete conclusions, but it is clear that these have serious consequences on both the understanding of oxidoreductase function and the ability to probe these enzymes as drug targets.