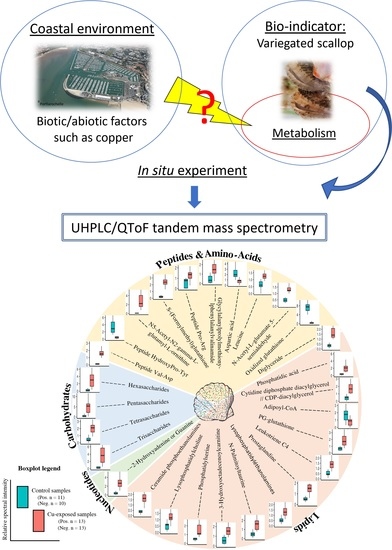
Untargeted Metabolomics Reveals a Complex Impact on Different Metabolic Pathways in Scallop Mimachlamys varia (Linnaeus, 1758) after Short-Term Exposure to Copper at Environmental Dose
Abstract
:1. Introduction
2. Results
2.1. LC/MS Data Processing and Analyses
2.2. Metabolite Modulation
3. Discussion
3.1. Energy Metabolism and Oxygen Transport
3.2. Inflammation, Oxidative Stress Defence and Lipid Metabolism
3.3. Osmoregulation, Reproduction
3.4. Peptide and Nucleotide Metabolism
4. Materials and Methods
4.1. Experimental Design and Sample Preparation
4.2. UHPLC/QToF MS Analysis of Samples
4.3. Statistical Analysis
4.4. Metabolite Annotation
- -
- Score 1, identification using a standard (same retention times, m/z and fragments).
- -
- Score 2a, annotation using fragmentation data from databases such as HMDB, LipidMaps, Metlin, with a spectrum-structure match unambiguous.
- -
- Score 2b, the fragments obtained match completely with the proposed structure, which excludes other possibilities, but the data are not completely available in the databases.
- -
- Score 3, proposed annotation of one or more isomeric molecules without the possibility of distinguishing between them because few or no fragments were obtained, or the fragments were common to the different positional isomers.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballabio, C.; Panagos, P.; Lugato, E.; Huang, J.H.; Orgiazzi, A.; Jones, A.; Fernández-Ugalde, O.; Borrelli, P.; Montanarella, L. Copper distribution in European topsoils: An assessment based on LUCAS soil survey. Sci. Total Environ. 2018, 636, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Weichenthal, S.; Kwong, J.C.; Burnett, R.T.; Hatzopoulou, M.; Jerrett, M.; van Donkelaar, A.; Bai, L.; Martin, R.V.; Copes, R.; et al. A population-based cohort study of respiratory disease and long-term exposure to iron and copper in fine particulate air pollution and their combined impact on reactive oxygen species generation in human lungs. Environ. Sci. Technol. 2021, 55, 3807–3818. [Google Scholar] [CrossRef] [PubMed]
- Blossom, N. Copper in the Ocean Environment. Am. Chemet Corp. 2015. Available online: https://www.chemet.com/assets/1/6/Copper_and_the_Ocean_Environment.pdf (accessed on 10 December 2021).
- Malhotra, N.; Ger, T.R.; Uapipatanakul, B.; Huang, J.C.; Chen, K.H.C.; Hsiao, C. Der Review of copper and copper nanoparticle toxicity in fish. Nanomaterials 2020, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Pandita, S.; Singh Sidhu, G.P.; Sharma, A.; Khanna, K.; Kaur, P.; Bali, A.S.; Setia, R. Copper bioavailability, uptake, toxicity and tolerance in plants: A comprehensive review. Chemosphere 2021, 262, 127810. [Google Scholar] [CrossRef] [PubMed]
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 2020, 114, 377–390. [Google Scholar] [CrossRef]
- Olivares, M.; Uauy, R. Copper as an essential nutrient. Am. J. Clin. Nutr. 1996, 63, 791S–796S. [Google Scholar] [CrossRef] [PubMed]
- Cairo, G.; Bernuzzi, F.; Recalcati, S. A precious metal: Iron, an essential nutrient for all cells. Genes Nutr. 2006, 1, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, J.C. Zinc: An essential but elusive nutrient. Am. J. Clin. Nutr. 2011, 94, 679S–684S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.V.; Alfaro, A.C.; Merien, F.; Lulijwa, R.; Young, T. Copper-induced immunomodulation in mussel (Perna canaliculus) haemocytes. Metallomics 2018, 10, 965–978. [Google Scholar] [CrossRef]
- Ory, P.; Hamani, V.; Bodet, P.E.; Murillo, L.; Graber, M. The variegated scallop, Mimachlamys varia, undergoes alterations in several of its metabolic pathways under short-term zinc exposure. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 37, 100779. [Google Scholar] [CrossRef]
- Kamunde, C.; Wood, C. Environmental chemistry, physiological homeostasis, toxicology, and environmental regulation of copper, an essential element in freshwater. Australas. J. Ecotoxicol. 2004, 10, 1–20. [Google Scholar]
- Van Holde, K.E.; Miller, K.I.; Decker, H. Hemocyanins and Invertebrate Evolution. J. Biol. Chem. 2001, 276, 15563–15566. [Google Scholar] [CrossRef] [Green Version]
- Gianazza, E.; Eberini, I.; Palazzolo, L.; Miller, I. Hemolymph proteins: An overview across marine arthropods and molluscs. J. Proteomics 2021, 245, 104294. [Google Scholar] [CrossRef]
- Vieira, L.R.; Gravato, C.; Soares, A.M.V.M.; Morgado, F.; Guilhermino, L. Acute effects of copper and mercury on the estuarine fish Pomatoschistus microps: Linking biomarkers to behaviour. Chemosphere. 2009, 76, 1416–1427. [Google Scholar] [CrossRef]
- Eklund, B.; Eklund, D. Pleasure boatyard soils are often highly contaminated. Environ. Manag. 2014, 53, 930–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brignon, J.M.; Gouzy, A. Données technico-économiques sur les substances chimiques en France: Cuivre, composés et alliages. INERIS 2014, 91, DRC-14-136881-02236A. Available online: http://www.ineris.fr/substances/fr/ (accessed on 10 December 2021).
- Huguet, J.R.; Brenon, I.; Coulombier, T. Characterisation of the water renewal in a macro-tidal marina using several transport timescales. Water 2019, 11, 2050. [Google Scholar] [CrossRef] [Green Version]
- Sim, V.X.Y.; Dafforn, K.A.; Simpson, S.L.; Kelaher, B.P.; Johnston, E.L. Sediment contaminants and infauna associated with recreational boating structures in a multi-use marine park. PLoS ONE 2015, 10, e0130537. [Google Scholar] [CrossRef]
- Connell, S.D.; Glasby, T.M. Do urban structures influence local abundance and diversity of subtidal epibiota? A case study from Sydney Harbour, Australia. Mar. Environ. Res. 1999, 47, 373–387. [Google Scholar] [CrossRef]
- Glasby, T.M. Differences between subtidal epibiota on pier pilings and rocky reefs at Marinas in Sydney, Australia. Estuar. Coast. Shelf Sci. 1999, 48, 281–290. [Google Scholar] [CrossRef]
- Connell, S.D. Floating pontoons create novel habitats for subtidal epibiota. J. Exp. Mar. Bio. Ecol. 2000, 247, 183–194. [Google Scholar] [CrossRef]
- Bustamante, P.; Miramand, P. Evaluation of the variegated scallop Chlamys varia as a biomonitor of temporal trends of Cd, Cu, and Zn in the field. Environ. Pollut. 2005, 138, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Breitwieser, M.; Barbarin, M.; Plumejeaud-Perreau, C.; Dubillot, E.; Guyot, T.; Huet, V.; Churlaud, C.; Coulombier, T.; Brenon, I.; Fichet, D.; et al. Biomonitoring of Mimachlamys varia transplanted to areas impacted by human activities (La Rochelle Marina, France). Chemosphere 2020, 243, 125199. [Google Scholar] [CrossRef]
- Milinkovitch, T.; Bustamante, P.; Huet, V.; Reigner, A.; Churlaud, C.; Thomas-Guyon, H. In situ evaluation of oxidative stress and immunological parameters as ecotoxicological biomarkers in a novel sentinel species (Mimachlamys varia). Aquat. Toxicol. 2015, 161, 170–175. [Google Scholar] [CrossRef]
- Breitwieser, M.; Becquet, V.; Thomas-Guyon, H.; Pillet, V.; Sauriau, P.G.; Graber, M.; Viricel, A. Population structure and genetic diversity in the variegated scallop, Mimachlamys varia (Linnaeus, 1758), a novel bioindicator of chemical pollution on the French coastline. J. Molluscan Stud. 2018, 84, 417–425. [Google Scholar] [CrossRef]
- Breitwieser, M.; Viricel, A.; Graber, M.; Murillo, L.; Becquet, V.; Churlaud, C.; Fruitier-Arnaudin, I.; Huet, V.; Lacroix, C.; Pante, E.; et al. Short-term and long-term biological effects of chronic chemical contamination on natural populations of a marine bivalve. PLoS ONE. 2016, 11, e0150184. [Google Scholar] [CrossRef] [Green Version]
- Metian, M.; Bustamante, P.; Hédouin, L.; Oberhänsli, F.; Warnau, M. Delineation of heavy metal uptake pathways (seawater and food) in the variegated scallop Chlamys varia, using radiotracer techniques. Mar. Ecol. Prog. Ser. 2009, 375, 161–171. [Google Scholar] [CrossRef] [Green Version]
- Metian, M.; Warnau, M.; Oberhänsli, F.; Bustamante, P. Delineation of Pb contamination pathways in two Pectinidae: The variegated scallop Chlamys varia and the king scallop Pecten maximus. Sci. Total Environ. 2009, 407, 3503–3509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitwieser, M.; Vigneau, E.; Viricel, A.; Becquet, V.; Lacroix, C.; Erb, M.; Huet, V.; Churlaud, C.; Le Floch, S.; Guillot, B.; et al. What is the relationship between the bioaccumulation of chemical contaminants in the variegated scallop Mimachlamys varia and its health status? A study carried out on the French Atlantic coast using the Path ComDim model. Sci. Total Environ. 2018, 640, 662–670. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; You, L.; Zhou, D.; Wu, H.; Li, L.; Zhao, J.; Feng, J.; Yu, J. Metabolic responses in gills of Manila clam Ruditapes philippinarum exposed to copper using NMR-based metabolomics. Mar. Environ. Res. 2011, 72, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.Y.; Wang, W.X. A lipidomic approach to understand copper resilience in oyster Crassostrea hongkongensis. Aquat. Toxicol. 2018, 204, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, M.; Viricel, A.; Churlaud, C.; Guillot, B.; Martin, E.; Stenger, P.L.; Huet, V.; Fontanaud, A.; Thomas-Guyon, H. First data on three bivalve species exposed to an intra-harbour polymetallic contamination (La Rochelle, France). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 199, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I.M.; Frederich, M.; Bagwe, R.; Lannig, G.; Sukhotin, A. A Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar. Environ. Res. 2012, 79, 1–15. [Google Scholar] [CrossRef]
- Ory, P.; Bonnet, A.; Mondeguer, F.; Breitwieser, M.; Dubillot, E.; Graber, M. Metabolomics based on UHPLC-QToF- and APGC-QToF-MS reveals metabolic pathways reprogramming in response to tidal cycles in the sub-littoral species Mimachlamys varia exposed to aerial emergence. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; Wang, W.X. Copper-induced metabolic variation of oysters overwhelmed by salinity effects. Chemosphere. 2017, 174. [Google Scholar] [CrossRef] [PubMed]
- Bayne, B.L.; Bubel, A.; Gabbott, P.A.; Livingstone, D.R.; Lowe, D.M.; Moore, M.N. Glycogen utilization and gametogenesis in Mytilus edulis (L.). Mar. Biol. Letters 1982, 3, 89–105. [Google Scholar]
- Ruiz, C.; Martinez, D.; Mosquera, G.; Abad, M.; Sánchez, J.L. Seasonal variations in condition, reproductive activity and biochemical composition of the flat oyster, Ostrea edulis, from San Cibran (Galicia, Spain). Mar. Biol. 1992, 112, 67–74. [Google Scholar] [CrossRef]
- Nakayama, A.; Yamamoto, K.; Tabata, S. Identification of the Catalytic Residues of Bifunctional Glycogen Debranching Enzyme. J. Biol. Chem. 2001, 276, 2882428828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollen, M.; Keppens, S.; Stalmans, W. Specific features of glycogen metabolism in the liver. Biochem. J. 1998, 336, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, K.; Yokoyama, I.; Suda, M.; Hata, M.; Matsuda, K. Purification and properties of glycogen phosphorylase from the adductor muscle of the scallop, Patinopecten yessoensis. Comp. Biochem. Physiol. Part B Biochem. 1987, 87, 747–753. [Google Scholar] [CrossRef]
- Smits, M.; Artigaud, S.; Bernay, B.; Pichereau, V.; Bargelloni, L.; Paillard, C. A proteomic study of resistance to Brown Ring disease in the Manila clam, Ruditapes philippinarum. Fish Shellfish Immunol. 2020, 99, 641–653. [Google Scholar] [CrossRef]
- Canesi, L.; Ciacci, C.; Piccoli, G.; Stocchi, V.; Viarengo, A.; Gallo, G. In vitro and in vivo effects of heavy metals on mussel digestive gland hexokinase activity: The role of glutathione. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998, 120, 261–268. [Google Scholar] [CrossRef]
- Le Saux, A.; David, E.; Betoulle, S.; Bultelle, F.; Rocher, B.; Barjhoux, I.; Cosio, C. New insights into cellular impacts of metals in aquatic animals. Environments 2020, 7, 46. [Google Scholar] [CrossRef]
- Cappel, R.E.; Bremer, J.W.; Timmons, T.M.; Nelson, T.E.; Gilbert, H.F. Thiol/disulfide redox equilibrium between glutathione and glycogen debranching enzyme (amylo-1,6-glucosidase/4-α-glucanotransferase) from rabbit muscle. J. Biol. Chem. 1986, 261, 15385–15389. [Google Scholar] [CrossRef]
- Connor, K.M.; Gracey, A.Y. High-resolution analysis of metabolic cycles in the intertidal mussel Mytilus californianus. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2012, 30, R103–R1112. [Google Scholar] [CrossRef] [Green Version]
- Gracey, A.Y.; Connor, K. Transcriptional and metabolomic characterization of spontaneous metabolic cycles in Mytilus californianus under subtidal conditions. Mar. Genom. 2016, 30, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Matsui, T.; Gatsogiannis, C.; Tanaka, Y. Molluscan hemocyanin: Structure, evolution, and physiology. Biophys. Rev. 2018, 10, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, D.W.; Brouwer, M.; McKenna, S. Hemocyanin concentrations in marine crustaceans as a function of environmental conditions. Mar. Ecol. Prog. Ser. 1993, 93, 235–244. [Google Scholar] [CrossRef]
- Ferreira, N.G.C.; Saborano, R.; Morgado, R.; Cardoso, D.N.; Rocha, C.M.; Soares, A.M.V.M.; Loureiro, S.; Duarte, I.F. Metabolic responses of the isopod Porcellionides pruinosus to nickel exposure assessed by 1H NMR metabolomics. J. Proteom. 2016, 137, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Tymoczko, J.; Stryer, L. Biochemistry, 5th ed.; W.H.Freeman & Co.: New York, NY, USA, 2002. [Google Scholar]
- Spector, A.A.; Yorek, M.A. Membrane lipid composition and cellular function. J. Lipid Res. 1985, 26, 1015–1035. [Google Scholar] [CrossRef]
- Yèagle, P.L. Lipid regulation of cell membrane structure and function. FASEB J. 1989, 3, 1833–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Lee, M.; Fairn, G.D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293, 6230–6240. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Li, M.; Zhong, Z.; Chen, H.; Wang, X.; Wang, M.; Xu, Z.; Cao, L.; Lian, C.; Zhang, H.; et al. Biochemical and metabolic responses of the deep-sea mussel Bathymodiolus platifrons to cadmium and copper exposure. Aquat. Toxicol. 2021, 236, 105845. [Google Scholar] [CrossRef]
- Fokina, N.N.; Ruokolainen, T.R.; Nemova, N.N.; Bakhmet, I.N. Changes of blue mussels Mytilus edulis L. lipid composition under cadmium and copper toxic effect. Biol. Trace Elem. Res. 2013, 154. [Google Scholar] [CrossRef]
- Soudant, P.; Marty, Y.; Moal, J.; Samain, J.F. Separation of major polar lipids in Pecten maximus by high-performance liquid chromatography and subsequent determination of their fatty acids using gas chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1995, 673, 15–26. [Google Scholar] [CrossRef]
- Tabakaeva, O.V.; Tabakaev, A. V Phospholipids from Soft Tissues of the Bivalve Mollusk Anadara broughtonii. Chem. Nat. Compd. 2016, 52, 299–300. [Google Scholar] [CrossRef]
- Canesi, L.; Ciacci, C.; Bergami, E.; Monopoli, M.P.; Dawson, K.A.; Papa, S.; Canonico, B.; Corsi, I. Evidence for immunomodulation and apoptotic processes induced by cationic polystyrene nanoparticles in the hemocytes of the marine bivalve Mytilus. Mar. Environ. Res. 2015, 111, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Di Gennaro, A.; Haeggström, J.Z. The Leukotrienes: Immune-Modulating Lipid Mediators of Disease; Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 116, ISBN 9780123943002. [Google Scholar]
- Becher, U.M.; Ghanem, A.; Tiyerili, V.; Fürst, D.O.; Nickenig, G.; Mueller, C.F.H. Inhibition of leukotriene C4 action reduces oxidative stress and apoptosis in cardiomyocytes and impedes remodeling after myocardial injury. J. Mol. Cell. Cardiol. 2011, 50, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, E.E.; Chupin, V.; de Kruijff, B.; Burger, K.N.J. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003, 4, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Nigou, J.; Besra, G.S. Cytidine diphosphate-diacylglycerol synthesis in Mycobacterium smegmatis. Biochem. J. 2002, 367, 157–162. [Google Scholar] [CrossRef]
- Blunsom, N.J.; Cockcroft, S. CDP-Diacylglycerol Synthases (CDS): Gateway to Phosphatidylinositol and Cardiolipin Synthesis. Front. Cell Dev. Biol. 2020, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Waluk, D.P.; Sucharski, F.; Sipos, L.; Silberring, J.; Hunt, M.C. Reversible lysine acetylation regulates activity of human glycine N-acyltransferase-like 2 (hGLYATL2): Implications for production of glycine-conjugated signaling molecules. J. Biol. Chem. 2012, 287, 16158–16167. [Google Scholar] [CrossRef] [Green Version]
- Sajiki, J.; Taguchi, S. Phospholipid Metabolism in Bivalves and their Feed Plankton. Biosci. Biotechnol. Biochem. 1995, 59, 1113–1117. [Google Scholar] [CrossRef]
- Frisca, F.; Sabbadini, R.A.; Goldshmit, Y.; Pébay, A. Biological Effects of Lysophosphatidic Acid in the Nervous System; International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 296, ISBN 9780123943071. [Google Scholar]
- Panevska, A.; Skočaj, M.; Križaj, I.; Maček, P.; Sepčić, K. Ceramide phosphoethanolamine, an enigmatic cellular membrane sphingolipid. Biochim. Biophys. Acta-Biomembr. 2019, 1861, 1284–1292. [Google Scholar] [CrossRef]
- Kunduri, G.; Turner-Evans, D.; Konya, Y.; Izumi, Y.; Nagashima, K.; Lockett, S.; Holthuis, J.; Bamba, T.; Acharya, U.; Acharya, J.K. Defective cortex glia plasma membrane structure underlies light-induced epilepsy in cpes mutants. Proc. Natl. Acad. Sci. USA 2018, 115, E8919–E8928. [Google Scholar] [CrossRef] [Green Version]
- Conners, D.E.; Ringwood, A.H. Effects of glutathione depletion on copper cytotoxicity in oysters (Crassostrea virginica). Aquat. Toxicol. 2000, 50, 341–349. [Google Scholar] [CrossRef]
- Paul, A.J. Glutathione Conjugation as a Determinant in 1,2-Dihaloethane and alpha-Naphthylisothiocyanate Toxicity. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 2 December 1991. [Google Scholar]
- Di Costanzo, F.; Di Dato, V.; Ianora, A.; Romano, G. Prostaglandins in marine organisms: A review. Mar. Drugs 2019, 17, 428. [Google Scholar] [CrossRef] [Green Version]
- Saintsing, D.G.; Dietz, T.H. Modification of sodium transport in freshwater mussels by prostaglandins, cyclic AMP and 5-hydroxytryptamine: Effects of inhibitors of prostaglandin synthesis. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1983, 76, 285–290. [Google Scholar] [CrossRef]
- Freas, W.; Grollman, S. Ionic and osmotic influence on prostaglandin release from the gill tissue of a marine bivalve, Modiolus demissus. J. Exp. Biol. 1980, 84, 169185. [Google Scholar] [CrossRef]
- Dumas, T.; Bonnefille, B.; Gomez, E.; Boccard, J.; Castro, N.A.; Fenet, H.; Courant, F. Metabolomics approach reveals disruption of metabolic pathways in the marine bivalve Mytilus galloprovincialis exposed to a WWTP effluent extract. Sci. Total Environ. 2020, 712, 136551. [Google Scholar] [CrossRef]
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Watanabe, M.; Meyer, K.A.; Jackson, T.M.; Schock, T.B.; Johnson, W.E.; Bearden, D.W. Application of NMR-based metabolomics for environmental assessment in the Great Lakes using zebra mussel (Dreissena polymorpha). Metabolomics 2015, 11, 1302–1315. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, H.; Kasai, H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J. Biol. Chem. 1995, 270, 19446–19450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesquita, A.F.; Marques, S.M.; Marques, J.C.; Gonçalves, F.J.M.; Gonçalves, A.M.M. Copper sulphate impact on the antioxidant defence system of the marine bivalves Cerastoderma edule and Scrobicularia plana. Sci. Rep. 2019, 9, 16458. [Google Scholar] [CrossRef] [PubMed]
- Ravera, O. Monitoring of the aquatic environment by species accumulator of pollutants: A review. J. Limnol. 2001, 60, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Trevisan, R.; Mello, D.F.; Delapedra, G.; Silva, D.G.H.; Arl, M.; Danielli, N.M.; Metian, M.; Almeida, E.A.; Dafre, A.L. Gills as a glutathione-dependent metabolic barrier in Pacific oysters Crassostrea gigas: Absorption, metabolism and excretion of a model electrophile. Aquat. Toxicol. 2016, 173, 105–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondeguer, F.; Abadie, E.; Herve, F.; Bardouil, M.; Sechet, V.; Raimbault, V.; Berteaux, T.; Zendong, S.Z.; Palvadeau, H.; Amzil, Z.; et al. Pinnatoxines en lien avec l’espèce Vulcanodinium rugosum (II). 2015. Available online: http://archimer.ifremer.fr/doc/00285/39635/ (accessed on 10 December 2021).
- Van Der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]



| Metabolite | Mode | Retention (min) | Formula | Monoisotopic Mass (Da) | Adduct | Observed Mass (m/z) | Theoretical Mass (m/z) | Mass Error (ppm) | Score | Copper Effect | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbohydrate | Trisaccharides | Pos | 1.45 | C18H32O16 | 504.1690 | [M + H]+ | 505.1777 | 505.1763 | 2.8 | 2a |  × 8 × 8 |
| Tetrasaccharides | Pos/Neg | 1.46 /2.52 | C24H42O21 | 666.2219 | [M + H]+ | 689.2123/665.2133 | 667.2291 | 1.5/2 | 2a |  × 4 × 4 | |
| Pentasaccharides | Pos | 4.64 | C30H52O26 | 828.2747 | [M + H + K]2+ | 434.1180 | 434.1226 | 10.5 | 2a |  × 10 × 10 | |
| Hexasaccharides | Pos | 5.72 | C36H62O31 | 990.3275 | [M + H + K]2+ | 515.1465 | 515.1490 | 4.9 | 2a |  × 10 × 10 | |
| Peptide & AAC | Peptide Val-Asp | Pos | 3.28 | C9H16N2O5 | 232.1059 | [M + H]+ | 233.1136 | 233.1132 | 1.6 | 2a |  × 8 × 8 |
| Peptide HydroxyPro-Tyr | Pos | 4.16 | C14H18N2O5 | 294.1216 | [M + H]+ | 333.0862 | 333.0847 | 4.5 | 2a |  × 6 × 6 | |
| N5-Acetyl-N2-gamma-l-glutamyl-l-ornithine | Pos | 4.64 | C12H21N3O6 | 303.1430 | [M + H]+ | 304.1507 | 304.1503 | 1.3 | 2a |  × 31 × 31 | |
| S-(Formylmethyl)glutathione | Pos | 6.39 | C12H19N3O7S | 349.0944 | [M+Na]+ | 372.0856 | 372.0836 | 5.2 | 2b |  × 10 × 10 | |
| Peptide Pro-Arg | Pos | 8.06 | C11H21N5O3 | 271.1644 | [M + H]+ | 310.1288 | 310.1276 | 4.0 | 2a |  × 2 × 2 | |
| Glycylalanylprolylmethionylphenylalanylvalinamide | Pos/Neg | 8.27 | C29H45N7O6S | 619.3152 | [M+2H]2+/ [M − H]− | 310.6649/618.3034 | 310.6649/618.3079 | 0/7.3 | 2b |  × 3 × 3 | |
| Aspartic acid | Neg | 1.11 | C4H7NO4 | 133.0375 | [M − H]− | 132.0295 | 132.0302 | 5.5 | 1 |  × 3 × 3 | |
| Leucine | Neg | 3.3 | C6H13NO2 | 131.0946 | [2M − 2H + Na]− | 283.1629 | 283.1634 | 1.7 | 1 |  × 2 × 2 | |
| N-Acetyl-l-glutamate 5-semialdehyde | Neg | 5.74 | C7H11NO4 | 173.0688 | [M − H]− | 172.0608 | 172.0615 | 4.3 | 2a |  × 2 × 2 | |
| Oxidized glutathione | Neg | 6.12 | C20H32N6O12S2 | 612.152 | [M − H]− | 611.1436 | 611.1447 | 1.8 | 2a |  × 13 × 13 | |
| Lipid | Diglyceride (9M5/9D3/0:0) or (11M3/9D3/0:0) or (9D3/11M3/0:0) or (9D3/9M5/0:0) | Pos | 6.49 | C40H66O7 | 658.4809 | [M + H + K]2+ | 349.2269 | 349.2256 | 3.8 | 3 |  × 5 × 5 |
| Phosphatidic acid | Pos | 7.11 | C41H69O8P | 720.4730 | [M + H + K]2+ | 380.2225 | 380.2217 | 2.1 | 3 |  × 5 × 5 | |
| Cytidine diphosphate diacylglycerol or CDP-diacylglycerol (CDP-DG) | Pos | 7.48 | C45H83N3O15P2 | 967.5299 | [M+H+Na]2+ | 495.7663 | 495.7632 | 6.3 | 3 |  × 4 × 4 | |
| Adipoyl-CoA | Neg | 6.81 | C27H44N7O19P3S | 895.1626 | [M − 2H]2− | 446.5629 | 446.5740 | 24.8 | 2a |  × 2 × 2 | |
| S-(PGA1) or S-(9-deoxy-D12-PGD2) or S-(11-OH-9-deoxy-D9,12-PGD2) or S-(9-deoxy-delta9,12-PGD2) or S-(PGJ2) or S-(PGA2)-glutathione | Neg | 8.58 | C30H47N3O10S | 641.2982 | [M − H]− | 640.2889 | 640.2909 | 3.1 | 3 |  × 3 × 3 | |
| Leukotriene C4 or 11-trans-Leukotriene C4 | Neg | 10.3 | C30H47N3O9S | 625.3033 | [M − H]− | 624.2950 | 624.2960 | 1.6 | 3 |  × 2 × 2 | |
| Prostaglandin D1, E1, F2 or H1 or as 8-isoprostaglandin F2 or E1 | Neg | 12.88 | C20H34O5 | 354.2406 | [M − H]− | 353.2325 | 353.2333 | 2.3 | 3 |  × 3 × 3 | |
| Lysophosphatidyléthanolamines (lysoPE) (P-16:0/0:0) and (0:0/18:2(9Z,12Z)) | Neg | 17.31/ 17.36 | C21H44NO(6/7)P | 437.2906/477.2855 | [M − H]− | 436.2816/476.2769 | 436.2833/476.2782 | 4/2.8 | 3 |  × 2 × 2 | |
| N-Palmitoyltaurine | Neg | 18.43 | C18H37NO4S | 363.2443 | [M − H]− | 362.2360 | 362.2371 | 3.1 | 2a |  × 3 × 3 | |
| 3-Hydroxyoctadecenoylcarnitine | Neg | 18.7 | C25H47NO5 | 441.3454 | [M + K − 2H]− | 478.295 | 478.294 | 2.1 | 3 |  × 3 × 3 | |
| Phosphatidylserine 20:5(5Z,8Z,11Z,14Z,17Z) or 18:3(9Z,12Z,15Z) or 18:4(6Z,9Z,12Z,15Z) or 20:4(8Z,11Z,14Z,17Z) | Neg | 19.51 | C44H70NO10P | 824.4495/803.4737 | [M + Na − 2H]−/[M − H]− | 824.4495/802.4676 | 824.4484/802.4665 | 1.3/1.4 | 3 |  × 3 × 3 | |
| Lysophosphatidylcholine (LysoPC) (18:2(9Z,12Z)/0:0) or (0:0/18:2(9Z,12Z)) | Neg | 20.54 | C26H50NO7P | 519.3325 | [M − H]− | 518.3237 | 518.3252 | 2.9 | 3 |  × 2 × 2 | |
| Ceramide phosphoethanolamines (PE-Cer) (d15:2(4E,6E)/20:0(2OH)) | Neg | 20.63 | C37H73N2O7P | 688.5155 | [M − H]−/[M + K − 2H]− | 687.5066 | 687.5083 | 2.5 | 3 |  × 4 × 4 | |
| Nucleotide | 2-Hydroxyadenine or Guanine | Pos | 4.52 | C5H5N5O | 151.0494 | [M + H]+ | 152.057 | 152.0567 | 2.0 | 2b/3 |  × 7 × 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamani, V.; Ory, P.; Bodet, P.-E.; Murillo, L.; Graber, M. Untargeted Metabolomics Reveals a Complex Impact on Different Metabolic Pathways in Scallop Mimachlamys varia (Linnaeus, 1758) after Short-Term Exposure to Copper at Environmental Dose. Metabolites 2021, 11, 862. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11120862
Hamani V, Ory P, Bodet P-E, Murillo L, Graber M. Untargeted Metabolomics Reveals a Complex Impact on Different Metabolic Pathways in Scallop Mimachlamys varia (Linnaeus, 1758) after Short-Term Exposure to Copper at Environmental Dose. Metabolites. 2021; 11(12):862. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11120862
Chicago/Turabian StyleHamani, Vincent, Pascaline Ory, Pierre-Edouard Bodet, Laurence Murillo, and Marianne Graber. 2021. "Untargeted Metabolomics Reveals a Complex Impact on Different Metabolic Pathways in Scallop Mimachlamys varia (Linnaeus, 1758) after Short-Term Exposure to Copper at Environmental Dose" Metabolites 11, no. 12: 862. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11120862