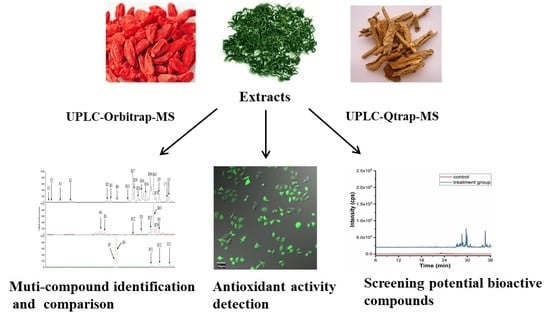
Comparative Study of the Chemical Constituents and Bioactivities of the Extracts from Fruits, Leaves and Root Barks of Lycium barbarum
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Identification of 5,6-dihydrosolasonine from Fruits
2.2. Multi-Component Analysis of Extracts by UPLC-HR-MS
2.3. Quantitative Analysis of Seven Compounds in the Fruits, Leaves and Root Barks
2.4. Antioxidative Activity Assays
2.5. Protective Effects of Extracts on H2O2-Induced Oxidative Stress in Cells
2.6. In Vitro Assays of Cytotoxicity
2.7. Potential Active Compounds Assay in Cells
3. Materials and Methods
3.1. Materials and Reagents
3.2. Sample Preparation
3.3. Isolation and Identification of 5,6-dihydrosolasonine from Fruit of L. barbarum
3.4. Multiple Component Identification by UPLC-Orbitrap-MS
3.5. Compound Database Construction by UPLC-Qtrap-MS
3.6. Quantitative Analysis of Compounds in Extracts
3.7. In Vitro Antioxidative Assays
3.7.1. DPPH Radical Scavenging Activity
3.7.2. ABTS Radical Scavenging Assay
3.7.3. FRAP Assay
3.8. Reactive Oxygen Species Measurement in L02 Cell
3.9. Cytotoxicity Assay
3.10. Target Cell-Based Screening of Potential Active Compounds
3.11. Data Handling and Presentation/Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, K.; Sasaki, T.; Li, W.; Li, Q.; Wang, Y.; Asada, Y.; Kato, H.; Koike, K. Two novel steroidal alkaloid glycosides from the seeds of Lycium barbarum. Chem. Biodivers. 2011, 8, 2277–2284. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, B.; Ma, H.R.; Aisa, H.A. Two new sesquiterpenoid glycosides from the leaves of Lycium barbarum. J. Asian Nat. Prod. Res. 2016, 18, 871–877. [Google Scholar] [CrossRef]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010, 76, 7–19. [Google Scholar] [CrossRef]
- Yu, M.S.; Leung, S.K.; Lai, S.W.; Che, C.M.; Zee, S.Y.; So, K.F.; Yuen, W.H.; Chang, R.C. Neuroprotective effects of anti-aging oriental medicine Lycium barbarum against beta-amyloid peptide neurotoxicity. Exp. Gerontol. 2005, 40, 716–727. [Google Scholar] [CrossRef]
- Amagase, H.; Farnsworth, N.R. A review of botanical characteristics, phytochemistry, clinical relevance in efficacy and safety of Lycium barbarum fruit (Goji). Food Res. Int. 2011, 44, 1702–1717. [Google Scholar] [CrossRef]
- Dong, J.Z.; Gao, W.S.; Lu, D.Y.; Wang, Y. Simultaneous Extraction and Analysis of Four Polyphenols from Leaves of Lycium Barbarum L. J. Food Biochem. 2011, 35, 914–931. [Google Scholar] [CrossRef]
- Yang, Y.; An, Y.; Wang, W.; Du, N.; Zhang, J.; Feng, Z.; Jiang, J.; Zhang, P. Nine compounds from the root bark of Lycium chinense and their anti-inflammatory activitieslammatory activitiesretain. Acta Pharm. Sin. B 2017, 7, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guan, S.; Sun, J.; Liu, T.; Chen, P.; Feng, R.; Chen, X.; Wu, W.; Yang, M.; Guo, D.A. Characterization and profiling of phenolic amides from Cortex Lycii by ultra-high performance liquid chromatography coupled with LTQ-Orbitrap mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 581–595. [Google Scholar] [CrossRef]
- Qian, D.; Zhao, Y.; Yang, G.; Huang, L. Systematic Review of Chemical Constituents in the Genus Lycium (Solanaceae). Molecules 2017, 22, 911. [Google Scholar] [CrossRef]
- Zhang, J.X.; Guan, S.H.; Feng, R.H.; Wang, Y.; Wu, Z.Y.; Zhang, Y.B.; Chen, X.H.; Bi, K.S.; Guo, D.A. Neolignanamides, lignanamides, and other phenolic compounds from the root bark of Lycium chinense. J. Nat. Prod. 2013, 76, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Mocan, A.; Vlase, L.; Vodnar, D.C.; Bischin, C.; Hanganu, D.; Gheldiu, A.M.; Oprean, R.; Silaghi-Dumitrescu, R.; Crisan, G. Polyphenolic content, antioxidant and antimicrobial activities of Lycium barbarum L. and Lycium chinense Mill. leaves. Molecules 2014, 19, 10056–10073. [Google Scholar] [CrossRef]
- Panidthananon, W.; Chaowasku, T.; Sritularak, B.; Likhitwitayawuid, K. A New Benzophenone C-Glucoside and Other Constituents of Pseuduvaria fragrans and Their alpha-Glucosidase Inhibitory Activity. Molecules 2018, 23, 1600. [Google Scholar] [CrossRef]
- Inbaraj, B.S.; Lu, H.; Kao, T.H.; Chen, B.H. Simultaneous determination of phenolic acids and flavonoids in Lycium barbarum Linnaeus by HPLC-DAD-ESI-MS. J. Pharm. Biomed. Anal. 2010, 51, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Mocan, A.; Vlase, L.; Raita, O.; Hanganu, D.; Paltinean, R.; Dezsi, S.; Gheldiu, A.M.; Oprean, R.; Crisan, G. Comparative studies on antioxidant activity and polyphenolic content of Lycium barbarum L. and Lycium chinense Mill. leaves. Pak. J. Pharm. Sci. 2015, 28, 1511–1515. [Google Scholar]
- Ren, W.; Han, L.; Luo, M.; Bian, B.; Guan, M.; Yang, H.; Han, C.; Li, N.; Li, T.; Li, S.; et al. Multi-component identification and target cell-based screening of potential bioactive compounds in toad venom by UPLC coupled with high-resolution LTQ-Orbitrap MS and high-sensitivity Qtrap MS. Anal. Bioanal. Chem. 2018, 410, 4419–4435. [Google Scholar] [CrossRef] [PubMed]
- Patras, M.A.; Jaiswal, R.; McDougall, G.J.; Kuhnert, N. Profiling and Quantification of Regioisomeric Caffeoyl Glucoses in Berry Fruits. J. Agric. Food Chem. 2018, 66, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Mocan, A.; Moldovan, C.; Zengin, G.; Bender, O.; Locatelli, M.; Simirgiotis, M.; Atalay, A.; Vodnar, D.C.; Rohn, S.; Crisan, G. UHPLC-QTOF-MS analysis of bioactive constituents from two Romanian Goji (Lycium barbarum L.) berries cultivars and their antioxidant, enzyme inhibitory, and real-time cytotoxicological evaluation. Food Chem. Toxicol. 2018, 115, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; Dai, D.; Li, L.; Wei, J.; Yang, H.; Li, S.; Zhang, Y.; Lin, Y.; Xiong, S.; Zhao, Z. Comprehensive qualification and quantification of triacylglycerols with specific fatty acid chain composition in horse adipose tissue, human plasma and liver tissue. Talanta 2017, 172, 206–214. [Google Scholar] [CrossRef]
- Lelario, F.; Labella, C.; Napolitano, G.; Scrano, L.; Bufo, S.A. Fragmentation study of major spirosolane-type glycoalkaloids by collision-induced dissociation linear ion trap and infrared multiphoton dissociation Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 2395–2406. [Google Scholar] [CrossRef]
- Lee, Y.-Y.; Hashimoto, F.; Yahara, S.; Nohara, T.; Yoshida, N. Study on the solanacenous plants. Part 29. Steroidal Glycosides from Solanum dulcamara. Chem. Pharm. Bull. 1994, 42, 707–709. [Google Scholar] [CrossRef]
- Matsushita, S.; Yanai, Y.; Fusyuku, A.; Ikeda, T.; Ono, M.; Nohara, T. Distinction of absolute configuration at C-22 of C-23-hydroxyspirostane and C-23-hydroxyspirosolane glycosides. Chem. Pharm. Bull. 2007, 55, 1079–1081. [Google Scholar] [CrossRef]
- Al Sinani, S.S.S.; Eltayeb, E.A. The steroidal glycoalkaloids solamargine and solasonine in Solanum plants. South Afr. J. Bot. 2017, 112, 253–269. [Google Scholar] [CrossRef]
- Trivedi, P.; Pundarikakshudu, K.J.C. Novel TLC Densitometric Method for Quantification of Solasodine in Various Solanum Species, Market Samples and Formulations. Chromatographia 2007, 65, 239–243. [Google Scholar] [CrossRef]
- Dinan, L.; Harmatha, J.; Lafont, R. Chromatographic procedures for the isolation of plant steroids. J. Chromatogr. A 2001, 935, 105–123. [Google Scholar] [CrossRef]
- Milner, S.E.; Brunton, N.P.; Jones, P.W.; O’Brien, N.M.; Collins, S.G.; Maguire, A.R. Bioactivities of glycoalkaloids and their aglycones from Solanum species. J. Agric. Food Chem. 2011, 59, 3454–3484. [Google Scholar] [CrossRef] [PubMed]
- Weissenberg, M. Isolation of solasodine and other steroidal alkaloids and sapogenins by direct hydrolysis-extraction of Solanum plants or glycosides therefrom. Phytochemistry 2001, 58, 501–508. [Google Scholar] [CrossRef]
- Roddick, J.G.; Weissenberg, M.; Leonard, A.L. Membrane disruption and enzyme inhibition by naturally-occurring and modified chacotriose-containing Solanum steroidal glycoalkaloids. Phytochemistry 2001, 56, 603–610. [Google Scholar] [CrossRef]
- Zhou, Z.Q.; Xiao, J.; Fan, H.X.; Yu, Y.; He, R.R.; Feng, X.L.; Kurihara, H.; So, K.F.; Yao, X.S.; Gao, H. Polyphenols from wolfberry and their bioactivities. Food Chem. 2017, 214, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Wolfram, K.; Schmidt, J.; Wray, V.; Milkowski, C.; Schliemann, W.; Strack, D. Profiling of phenylpropanoids in transgenic low-sinapine oilseed rape (Brassica napus). Phytochemistry 2010, 71, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Funayama, S.; Zhang, G.-R.; Nozoe, S.; Kukoamine, B. A spermine alkaloid from Lycium chinense. Phytochemistry 1995, 38, 1529–1531. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, H.; Zhao, C.; Huang, Y.Q.; Tang, X.; Cheung, H.Y. Identification and Characterization of Kukoamine Metabolites by Multiple Ion Monitoring Triggered Enhanced Product Ion Scan Method with a Triple-Quadruple Linear Ion Trap Mass Spectrometer. J. Agric. Food Chem. 2015, 63, 10785–10790. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.Q.; Fan, H.X.; He, R.R.; Xiao, J.; Tsoi, B.; Lan, K.H.; Kurihara, H.; So, K.F.; Yao, X.S.; Gao, H. Lycibarbarspermidines A-O, New Dicaffeoylspermidine Derivatives from Wolfberry, with Activities against Alzheimer’s Disease and Oxidation. J. Agric. Food Chem. 2016, 64, 2223–2237. [Google Scholar] [CrossRef] [PubMed]
- Yossa Nzeuwa, I.B.; Xia, Y.; Qiao, Z.; Feng, F.; Bian, J.; Liu, W.; Qu, W. Comparison of the origin and phenolic contents of Lycium ruthenicum Murr. by high-performance liquid chromatography fingerprinting combined with quadrupole time-of-flight mass spectrometry and chemometrics. J. Sep. Sci. 2017, 40, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xu, F.; Ji, T.; Li, J. A New Spermidine from the Fruits of Lycium ruthenicum. Chem. Nat. Compounds 2014, 50, 880–883. [Google Scholar] [CrossRef]
- Narváez-Cuenca, C.-E.; Vincken, J.-P.; Gruppen, H. Identification and quantification of (dihydro) hydroxycinnamic acids and their conjugates in potato by UHPLC–DAD–ESI-MSn. Food Chem. 2012, 130, 730–738. [Google Scholar] [CrossRef]
- Seca, A.M.; Silva, A.M.; Silvestre, A.J.; Cavaleiro, J.A.; Domingues, F.M.; Pascoal-Neto, C. Lignanamides and other phenolic constituents from the bark of kenaf (Hibiscus cannabinus). Phytochemistry 2001, 58, 1219–1223. [Google Scholar] [CrossRef]
- Strehmel, N.; Bottcher, C.; Schmidt, S.; Scheel, D. Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 2014, 108, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, H.; Wang, Q.-H.; Yang, B.-Y.; Kuang, H.-X. A new feruloyl tyramine glycoside from the roots of Achyranthes bidentata. Chin. J. Nat. Med. 2012, 10, 16–19. [Google Scholar] [CrossRef]
- Gao, K.; Ma, D.; Cheng, Y.; Tian, X.; Lu, Y.; Du, X.; Tang, H.; Chen, J. Three New Dimers and Two Monomers of Phenolic Amides from the Fruits of Lycium barbarum and Their Antioxidant Activities. J. Agric. Food Chem. 2015, 63, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Jones, S.H.; Hecht, S.M. Phenolic acid amides: a new type of DNA strand scission agent from Piper caninum. Bioorg. Med. Chem. 2004, 12, 3885–3889. [Google Scholar] [CrossRef]
- Sun, J.; Gu, Y.F.; Su, X.Q.; Li, M.M.; Huo, H.X.; Zhang, J.; Zeng, K.W.; Zhang, Q.; Zhao, Y.F.; Li, J.; et al. Anti-inflammatory lignanamides from the roots of Solanum melongena L. Fitoterapia 2014, 98, 110–116. [Google Scholar] [CrossRef]
- Mocan, A.; Zengin, G.; Simirgiotis, M.; Schafberg, M.; Mollica, A.; Vodnar, D.C.; Crisan, G.; Rohn, S. Functional constituents of wild and cultivated Goji (L. barbarum L.) leaves: Phytochemical characterization, biological profile, and computational studies. J. Enzyme. Inhib. Med. Chem. 2017, 32, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.B.; Ryu, J.; Cho, Y.; Choi, S.Z.; Son, M.; Sung, S.H. Combined Application of UHPLC-QTOF/MS, HPLC-ELSD and (1) H-NMR Spectroscopy for Quality Assessment of DA-9801, A Standardised Dioscorea Extract. Phytochem. Anal. 2017, 28, 185–194. [Google Scholar] [CrossRef]
- Challinor, V.L.; Parsons, P.G.; Chap, S.; White, E.F.; Blanchfield, J.T.; Lehmann, R.P.; De Voss, J.J. Steroidal saponins from the roots of Smilax sp.: structure and bioactivity. Steroids 2012, 77, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Su, X.; Huang, Z.; Su, L.; Li, L.; Lu, T. Analysis of Chemical Variations between Crude and Salt-Processed Anemarrhenae rhizoma Using Ultra-High-Performance Liquid Chromatography-Mass Spectrometry Methods. Molecules 2017, 23, 23. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Escobar, C.A.; Priego-Capote, F.; Luque de Castro, M.D. Characterization of lemon (Citrus limon) polar extract by liquid chromatography-tandem mass spectrometry in high resolution mode. J. Mass Spectrom. 2015, 50, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Kokotkiewicz, A.; Migas, P.; Stefanowicz, J.; Luczkiewicz, M.; Krauze-Baranowska, M. Densitometric TLC analysis for the control of tropane and steroidal alkaloids in Lycium barbarum. Food Chem. 2017, 221, 535–540. [Google Scholar] [CrossRef]
- Shanker, K.; Gupta, S.; Srivastava, P.; Srivastava, S.K.; Singh, S.C.; Gupta, M.M. Simultaneous determination of three steroidal glycoalkaloids in Solanum xanthocarpum by high performance thin layer chromatography. J. Pharm. Biomed. Anal. 2011, 54, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Affes, M.; Fakhfakh, J.; Daoud, I.; Brieudes, V.; Halabalaki, M.; El Feki, A.; Allouche, N. UHPLC/HR-ESI-MS/MS Profiling of Phenolics from Tunisian Lycium arabicum Boiss. Antioxidant and Anti-lipase Activities’ Evaluation. Chem. Biodivers. 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Cai, Y.; Yan, J.; Sun, M.; Corke, H. Hypoglycemic and hypolipidemic effects and antioxidant activity of fruit extracts from Lycium barbarum. Life Sci. 2004, 76, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, J.; Chen, B.; Xie, H.; Chen, D. Antioxidant and Cytoprotective Effects of Kukoamines A and B: Comparison and Positional Isomeric Effect. Molecules 2018, 23, 973. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry and anticarcinogenic mechanisms of glycoalkaloids produced by eggplants, potatoes, and tomatoes. J. Agric. Food Chem. 2015, 63, 3323–3337. [Google Scholar] [CrossRef]
- Hong, M.; Wang, X.Z.; Wang, L.; Hua, Y.Q.; Wen, H.M.; Duan, J.A. Screening of immunomodulatory components in Yu-ping-feng-san using splenocyte binding and HPLC. J. Pharm. Biomed. Anal. 2011, 54, 87–93. [Google Scholar] [CrossRef]
- Song, Y.; Song, Q.; Liu, Y.; Li, J.; Wan, J.B.; Wang, Y.; Jiang, Y.; Tu, P. Integrated work-flow for quantitative metabolome profiling of plants, Peucedani Radix as a case. Anal. Chim. Acta 2017, 953, 40–47. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.Q.; Xiao, H.; Zhang, J.; Liu, A. Simultaneous Qualitative Assessment and Quantitative Analysis of Metabolites (Phenolics, Nucleosides and Amino Acids) from the Roots of Fresh Gastrodia elata Using UPLC-ESI-Triple Quadrupole Ion MS and ESI- Linear Ion Trap High-Resolution MS. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Li, T.; Liu, C.; Liu, W.; Liu, J. Comparison of bioactivities and phenolic composition of Choerospondias axillaris peels and fleshes. J. Sci. Food Agric. 2016, 96, 2462–2471. [Google Scholar] [CrossRef]
- Can-Cauich, C.A.; Sauri-Duch, E.; Moo-Huchin, V.M.; Betancur-Ancona, D.; Cuevas-Glory, L.F. Effect of extraction method and specie on the content of bioactive compounds and antioxidant activity of pumpkin oil from Yucatan, Mexico. Food Chem. 2019, 285, 186–193. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Liu, R.H. Cellular antioxidant activity (CAA) assay for assessing antioxidants, foods, and dietary supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef]
Sample Availability: Not available. |






| No | RT 1 | Formula | [M + H]+ | [M − H]− | ppm | MS/MS Fragments 2 | Identification | Ref. 3 | F 4 | L 5 | R 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 8.70 | C21H29O13 | 487.1485 | 1.78 | 324.8671; 303.7701; 163.0403; 145.0296; 119.0503 | Lycibarbarphenylpropanoid A isomer | [28] | ✓ | |||
| A2 | 9.72 | C21H29O13 | 487.1492 | 1.22 | 398.1759; 229.0514; 163.0381; 145.0275; 119.0503 | Lycibarbarphenylpropanoid A isomer | [28] | ✓ | |||
| A3 | 10.44 | C22H30O14 | 517.1583 | 0.72 | 334.8634; 235.0619; 193.0510; 175.0403; 160.0169; 134.0375 | Lycibarbarphenylpropanoid C isomer | [28] | ✓ | |||
| A4 | 10.47 | C15H16O8 | 325.0937 | 0.23 | 298.8872; 163.0398; 145.0276; 119.0497 | p-coumaric acid O-glycosides | [16] | ✓ | ✓ | ||
| A5 | 15.45 | C16H18O9 | 355.1050 | 353.0854 | 1.34 | 308.9027; 285.0117; 181.0513; 163.0402; 145.0296; | chlorogenic acid isomer | [28] | ✓ | ||
| A6 | 16.16 | C22H30O14 | 517.1594 | 1.58 | 354.5810; 259.0620; 193.0510; 175.0403; 160.0169; 134.0375 | Lycibarbarphenylpropanoid C isomer | [28] | ✓ | |||
| A7 | 16.88 | C16H18O9 | 355.1050 | 353.0854 | 1.55 | 163.0395; 145.0289; 135.0445; 117.0339; 89.0390; | chlorogenic acid * | ✓ | |||
| A8 | 17.32 | C23H32O13 | 515.1417 | 1.00 | 395.0988; 353.0877; 274.9858; 191.0557; 161.0242 | Lycibarbarphenylpropanoid F isomer | [28] | ✓ | ✓ | ||
| A9 | 18.78 | C23H32O13 | 515.1422 | 1.24 | 323.0873; 274.9858; 191.0539; 161.0225 | Lycibarbarphenylpropanoid F isomer | [28] | ✓ | ✓ | ||
| A10 | 18.87 | C16H18O9 | 355.1008 | 353.0855 | 1.42 | 303.0228; 193.0512; 178.0276; 134.0375 | Scopolin * | ✓ | |||
| A11 | 18.92 | C27H36O18 | 647.1873 | 1.34 | 485.2170; 323.1666; 191.0319; 176.0087; 161.0431; 148.0142 | lycibarbarcoumarin A | [28] | ✓ | |||
| A12 | 20.42 | C16H18O9 | 355.1047 | 353.0854 | 1.34 | 285.0116; 193.0510; 163.0403; 145.0295; 123.1177 | chlorogenic acid isomer | [28] | ✓ | ||
| A13 | 30.04 | C17H22O10 | 385.1140 | 1.29 | 326.9597; 185.0199; 163.0381; 119.0486 | sinapate 4-O-β-glucopyranoside | [29] | ✓ | ✓ | ||
| B1 | 17.87 | C28H42O6N4 | 531.3170 | 529.3001 | 1.41 | 513.3074; 367.2724; 293.1855; 222.1123; 165.0546 | kukoamine B ismoer | [30] | ✓ | ||
| B2 | 18.48 | C40H62O16N4 | 855.4222 | 853.4034 | 1.38 | 693.3734; 529.3273; 455.2405; 384.1668; 293.1871; 222.1121; 165.0545 | 2Glu-[kukoamine] ismoer | ✓ | |||
| B3 | 19.30 | C40H62O16N4 | 855.4222 | 853.4034 | 1.38 | 693.3693; 531.3206; 455.2379; 384.1668; 293.1853; 222.1121; 165.0545 | 2Glu-[kukoamine] ismoer | ✓ | |||
| B4 | 19.48 | C46H72O21N4 | 1017.4749 | 1015.4559 | 1.31 | 855.4216; 617.2904; 455.2379; 384.1645; 222.1122; 165.0557 | 3Glu-[kukoamine] ismoer | ✓ | |||
| B5 | 19.50 | C28H42O6N4 | 531.3213 | 529.3011 | 1.41 | 513.3074; 367.2728; 293.1855; 222.1123; 165.0546 | kukoamine B ismoer | ✓ | |||
| B6 | 19.59 | C34H52O11N4 | 693.3698 | 691.3521 | 1.05 | 531.3168; 367.2730; 293.1855; 222.1122; 165.0560; 123.0440 | Glu-[kukoamine] ismoer | ✓ | |||
| B7 | 20.28 | C28H42O6N4 | 531.3165 | 529.3001 | 1.41 | 513.3103; 447.8046; 376.2692; 293.1879; 222.1140; 165.8764 | kukoamine A | [30] | ✓ | ||
| B8 | 20.60 | C46H72O21N4 | 1017.4747 | 1015.4559 | 1.49 | 855.4216; 617.2901; 455.2379; 384.1642; 222.1121; 165.0552 | 3Glu-[kukoamine] ismoer | ✓ | |||
| B9 | 20.65 | C28H42O6N4 | 531.3220 | 529.3001 | 1.41 | 513.3074; 367.2712; 310.2152; 293.1855; 222.1123; 165.0545 | kukoamine B * | ✓ | |||
| B10 | 21.02 | C28H42O6N4 | 531.3199 | 529.3003 | 1. 40 | 402.9773; 367.2716; 293.1880; 222.1141; 193.0510; 165.0553; 129.1395 | kukoamine B ismoer | [30] | ✓ | ||
| B11 | 21.73 | C28H40O6N4 | 529.3059 | 527.2831 | 1.13 | 472.2346; 367.2725; 293.1877; 222.1140; 163.0402 | Dihydrocaffeoyl quinonespermine ismoer 7 | [31] | ✓ | ||
| B12 | 23.31 | C28H40O6N4 | 529.3012 | 527.2847 | 1.72 | 511.2894; 455.2384; 384.1648; 293.1855; 220.0986; 163.0406 | Dihydrocaffeoyl quinonespermine ismoer | [31] | ✓ | ✓ | |
| B13 | 23.83 | C43H63O21N3 | 958.4016 | 956.3817 | 1.18 | 796.3486; 634.2960; 472.2396; 310.2119; 220.0966; 163.0388 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B14 | 24.41 | C37H51O16N3 | 794.3339 | 792.3145 | 0.36 | 632.2804; 470.2540; 382.1489; 220.0965; 163.0388 | [lycibarbarspermidine O] isomer | [32] | ✓ | ||
| B15 | 24.41 | C43H65O21N3 | 960.4161 | 958.3975 | 2.28 | 798.3622; 636.3112; 474.2588; 384.1645; 222.1122; 163.0402 | Glu-[lycibarbarspermidine M]ismoer | ✓ | |||
| B16 | 24.75 | C37H55O16N3 | 798.3640 | 796.3453 | 1.80 | 636.3071; 474.2589; 384.1644; 220.0965; 163.0388 | [lycibarbarspermidine M] isomer | [32] | ✓ | ||
| B17 | 24.78 | C37H53O16N3 | 796.3492 | 794.3301 | 180 | 634.2957; 472.2431; 310.2126; 220.0965; 163.0398 | [lycibarbarspermidine F] isomer | [32] | ✓ | ||
| B18 | 24.80 | C31H43O11N3 | 634.2959 | 632.2784 | 1.31 | 472.2431; 310.2122; 220.0966; 163.0390 | lycibarbarspermidine B isomer | [32] | ✓ | ||
| B19 | 24.94 | C43H63O21N3 | 958.4267 | 956.3817 | 1.82 | 796.3481; 634.2957; 472.2414; 310.2119; 220.0964; 163.0388 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B20 | 25.17 | C37H55O16N3 | 798.3640 | 796.3453 | 1.89 | 636.3117; 474.2589; 384.1644; 222.1121; 165.0545 | [lycibarbarspermidine M] isomer | [32] | ✓ | ||
| B21 | 25.62 | C28H42O5N4 | 515.3267 | 7.61 | 498.2995; 367.2717; 293.7878; 277.1928; 222.1139; 165.0556; 129.1395 | Dihydrocaffeoyl spermine derivative | ✓ | ||||
| B22 | 25.72 | C28H40O6N4 | 529.3054 | 527.2847 | 7.16 | 458.2317; 367.2724; 291.1721; 220.0988; 163.0401 | Dihydrocaffeoyl quinonespermine ismoer | [31] | ✓ | ✓ | |
| B23 | 25.72 | C28H40O6N4 | 529.3059 | 527.2847 | 1.72 | 511.2894; 393.2533; 384.1649; 291.1745; 220.2846; 163.0422 | Dihydrocaffeoyl quinonespermine ismoer | [31] | ✓ | ||
| B24 | 25.77 | C43H63O21N3 | 958.4016 | 956.3817 | 1.69 | 796.3489; 634.2968; 472.2392; 310.2118; 220.09656; 163.0383 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B25 | 26.02 | C37H55O16N3 | 798.3626 | 796.3453 | 3.63 | 636.3112; 474.2581; 384.1645; 222.1128; 165.0544 | [lycibarbarspermidine M] isomer | [32] | ✓ | ||
| B26 | 26.10 | C37H53O16N3 | 796.3409 | 794.3301 | 1.92 | 634.2957; 472.2431; 310.2121; 220.0965; 163.0390 | [lycibarbarspermidine F] isomer | [32] | ✓ | ||
| B27 | 26.36 | C43H65O21N3 | 960.4125 | 958.3969 | 3.30 | 798.3621; 636.3112; 474.2590; 384.1646; 222.1121; 163.0404 | Glu-[lycibarbarspermidine M]ismoer | ✓ | |||
| B28 | 26.41 | C43H63O21N3 | 958.4011 | 956.3817 | 1.63 | 796.3483; 634.2957; 472.2428; 398.1824; 310.2119; 220.0965; 163.0388 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B29 | 26.44 | C37H55O16N3 | 798.3609 | 796.3453 | 1.58 | 636.3071; 474.2585; 384.16455; 222.1122; 163.0385 | [lycibarbarspermidine M] isomer | ✓ | |||
| B30 | 26.79 | C43H63O21N3 | 958.4009 | 956.3817 | 1.82 | 796.3530; 634.2994; 472.2431; 382.1511; 310.2120; 220.0978; 163.0398 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B31 | 27.02 | C31H43O11N3 | 634.2964 | 632.2784 | 1.03 | 472.24500; 310.21201; 220.09660; 163.03981 | lycibarbarspermidine B isomer | [32] | ✓ | ||
| B32 | 27.03 | C31H41O11N3 | 632.2814 | 630.2632 | 0.07 | 470.2284; 382.1489; 308.1962; 220.0965; 163.0388 | lycibarbarspermidine N isomer | [32] | ✓ | ||
| B33 | 27.23 | C43H63O21N3 | 958.3870 | 956.3817 | 1.80 | 796.34717; 634.29457; 472.2424; 310.2124; 220.0964; 163.0383 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B34 | 27.42 | C43H65O21N3 | 960.4155 | 958.3969 | 9.00 | 798.3629; 636.3112; 474.2588; 384.1645; 222.1121; 165.0404 | Glu-[lycibarbarspermidine M] ismoer | ✓ | |||
| B35 | 27.51 | C31H45O11N3 | 636.3115 | 634.2941 | 2.00 | 474.2589; 384.1649; 222.1121; 165.0544 | lycibarbarspermidine J | ✓ | |||
| B36 | 27.73 | C31H41O11N3 | 632.2811 | 630.2632 | 0.50 | 470.2276; 382.1480; 308.1962; 220.0963; 163.0388 | lycibarbarspermidine N isomer | ✓ | |||
| B37 | 27.85 | C37H51O16N3 | 794.3337 | 792.3145 | 0.67 | 632.28010; 470.2540; 382.1489; 220.0965; 163.0388 | [lycibarbarspermidine O] isomer | ✓ | |||
| B38 | 27.91 | C43H63O21N3 | 958.3870 | 956.3817 | 1.22 | 796.3475; 634.2955; 472.2420; 310.2120; 220.09654; 163.0388 | Glu-[lycibarbarspermidine F] isomer | ✓ | |||
| B39 | 28.04 | C37H53O16N3 | 796.3495 | 794.3301 | 1.39 | 634.2959; 472.2431; 310.2122; 220.0966; 163.0391 | [lycibarbarspermidine F] isomer | [32] | ✓ | ||
| B40 | 28.16 | C43H61O21N3 | 956.3857 | 954.3660 | 1.48 | 794.3328; 632.2972; 470.2267; 220.0965; 163.0387 | Glu-[lycibarbarspermidine O] ismoer | ✓ | |||
| B41 | 28.30 | C43H65O21N3 | 960.4146 | 958.3969 | 3.39 | 798.3626; 636.3109; 474.2587; 384.1645; 222.1122; 165.0404 | Glu-[lycibarbarspermidine M]ismoer | ✓ | |||
| B42 | 28.57 | C31H43O11N3 | 634.2960 | 632.2784 | 1.69 | 472.2424; 382.1488; 310.2118; 220.0969; 163.0388 | lycibarbarspermidine B isomer | [32] | ✓ | ||
| B43 | 28.64 | C29H44O6N4 | 545.3377 | 543.3162 | 7.85 | 527.3255; 432.0255; 322.2652; 293.1878; 236.1295; 222.1139; 129.1396 | Dihydrocaffeoyl spermine derivative | ✓ | |||
| B44 | 28.77 | C41H57O20N3 | 912.3596 | 910.3403 | 1.38 | 750.3066; 634.2957; 472.2431; 310.2121; 220.0965; 163.0398 | Dihydrocaffeoyl spermidine derivative | ✓ | |||
| B45 | 28.83 | C29H44O6N4 | 545.3377 | 543.3162 | 7.97 | 527.3264; 381.2884; 307.4034; 293.1878; 222.1139; 165.14386; 129.1396 | Dihydrocaffeoyl spermine derivative | ✓ | |||
| B46 | 29.06 | C49H73O26N3 | 1120.4537 | 1118.4336 | 1.58 | 958.3557; 796.3481; 634.2957; 310.2118; 220.0965; 163.0402 | 2Glu-[lycibarbarspermidine F] ismoer | ✓ | |||
| B47 | 29.11 | C41H57O20N3 | 912.3597 | 910.3403 | 1.25 | 750.3068; 634.2960; 472.2431; 310.2121; 220.0965; 163.0398 | Dihydrocaffeoyl spermidine derivative | ✓ | |||
| B48 | 29.16 | C31H43O11N3 | 634.2959 | 632.2780 | 1.79 | 617.2727; 558.1018; 472.2414; 310.2120; 220.0966; 163.0388 | lycibarbarspermidine B isomer | [32] | ✓ | ✓ | |
| B49 | 29.23 | C31H41O11N3 | 632.2813 | 630.2630 | 0.12 | 604.2890; 587.2619; 470.2543; 382.1489; 308.1963; 220.0965; 163.0388 | lycibarbarspermidine N isomer | ✓ | |||
| B50 | 29.29 | C49H73O26N3 | 1120.4536 | 1118.4336 | 1.69 | 958.3558; 796.34814; 634.2957; 472.2396; 310.2118; 220.0965; 163.0412 | 2Glu-[lycibarbarspermidine F] ismoer | ✓ | |||
| B51 | 29.32 | C37H51O16N3 | 794.3336 | 792.3145 | 0.82 | 632.2830; 470.2540; 382.1489; 220.0966; 163.0389 | [lycibarbarspermidine O] isomer | ✓ | |||
| B52 | 29.40 | C25H35O6N3 | 474.2585 | 472.2435 | 2.79 | 457.2319; 310.2120; 222.1121; 165.0544; 123.0438 | N1-N10dihydrocaffeoyl spermidine | [31,33] | ✓ | ✓ | ✓ |
| B53 | 29.44 | C41H57O20N3 | 912.3593 | 910.3403 | 1.71 | 750.3066; 634.2957; 498.1598; 310.2121; 220.0965; 163.0398 | Dihydrocaffeoyl spermidine derivative | ✓ | |||
| B54 | 29.58 | C49H73O26N3 | 1120.4535 | 1118.4336 | 1.80 | 958.3557; 796.3481; 634.2957; 310.2115; 220.0965; 163.0412 | 2Glu-[lycibarbarspermidine F] ismoer | ✓ | |||
| B55 | 29.59 | C31H43O11N3 | 634.2960 | 632.2784 | 1.70 | 513.0655; 472.2426; 310.2116; 222.1120; 163.0388 | lycibarbarspermidine B isomer | ✓ | |||
| B56 | 29.60 | C31H41O11N3 | 632.2805 | 630.2630 | 1.42 | 496.2304; 470.2259; 382.1489; 308.1962; 220.0965; 163.0388 | lycibarbarspermidine N isomer | ✓ | |||
| B57 | 29.64 | C41H57O20N3 | 912.3593 | 910.3403 | 1.71 | 750.3065; 588.2751; 382.1488; 310.2121; 220.0964; 163.0398 | Dihydrocaffeoyl spermidine derivative | ✓ | |||
| B58 | 29.71 | C37H53O16N3 | 796.3480 | 794.3300 | 2.31 | 634.2957; 472.2431; 310.2121; 220.0965; 163.0390 | [lycibarbarspermidine F] isomer | ✓ | |||
| B59 | 29.71 | C37H51O16N3 | 794.3331 | 792.3145 | 1.44 | 632.2804; 470.2540; 308.1962; 220.0964; 163.0388 | [lycibarbarspermidine O] isomer | ✓ | |||
| B60 | 29.74 | C35H47O15N3 | 750.3065 | 748.2889 | 1.11 | 588.2747; 472.2429; 310.2117; 220.0964; 163.0387 | Glu-[lycibarbarspermidine B] ismoer | ✓ | |||
| B61 | 29.74 | C49H71O26N3 | 1118.4481 | 1116.4180 | 1.45 | 956.3852; 794.3327; 632.3016; 470.2233; 220.0965; 163.0388 | 2Glu-[lycibarbarspermidine O] isomer | ✓ | |||
| B62 | 29.88 | C35H49O15N3 | 750.3069 | 748.2889 | 1.48 | 588.2748; 472.2429; 310.2118; 220.0965; 163.0389 | Glu-[lycibarbarspermidine B] ismoer | ✓ | |||
| B63 | 30.03 | C25H33O6N3 | 472.2389 | 470.2264 | 1.64 | 455.2168; 310.2119; 220.0965; 163.0388; 112.1121 | N1–caffeoyl, N3-dihydrocaffeoyl spermidine | [34] | ✓ | ✓ | ✓ |
| B64 | 30.13 | C31H41O11N3 | 632.2804 | 630.2628 | 1.57 | 470.2497; 382.1489; 308.1962; 220.0965; 163.0388 | lycibarbarspermidine N isomer | ✓ | |||
| B65 | 30.14 | C28H41O8N3 | 548.3005 | 7.11 | 530.2393; 474.2629; 293.1877; 222.1940; 165.0555; 128.1080 | Dihydrocaffeoyl spermidine derivative | ✓ | ||||
| B66 | 30.14 | C34H28O12N4 | 705.3398 | 7.98 | 687.3212; 531.3211; 467.2042; 310.2160; 293.1826; 222.1139; 165.0559 | Dihydrocaffeoyl spermine derivative | ✓ | ||||
| B67 | 30.14 | C37H51O16N3 | 794.3336 | 792.3145 | 0.76 | 632.2804; 470.2540; 308.1963; 220.0965; 163.0388 | [lycibarbarspermidine O] isomer | ✓ | |||
| B68 | 30.19 | C30H46O6N4 | 559.3531 | 1.89 | 395.2663; 307.2033; 236.1295; 222.123; 165.0561 | Dihydrocaffeoyl spermine derivative | ✓ | ||||
| B69 | 30.19 | C28H40O7N3 | 530.2891 | 528.2696 | 1.68 | 474.2629; 310.2142; 293.1877; 222.1140; 165.0558 | Propionyl-dihydrocaffeoyl spermidine | ✓ | |||
| B70 | 30.56 | C25H31O6N3 | 470.2278 | 468.2115 | 1.54 | 308.1962; 291.1698; 234.1121; 220.0965; 163.0388 | N, N′-dicaffeoylspermidine | [33] | ✓ | ✓ | |
| B71 | 30.82 | C34H46O11N4 | 687.3308 | 685.3051 | 7.59 | 670.3035; 523.2778; 449.1942; 293.1878; 222.1139 | Dihydrocaffeoyl spermine derivative | ✓ | |||
| B72 | 31.45 | C37H50O9N4 | 695.3701 | 693.3464 | 1.28 | 678.3438; 531.3215; 457.2365; 293.1879; 222.1140; 165.0559 | N1,N4,N12-tris(dihydrocaffeoyl)spermine | [35] | ✓ | ||
| B73 | 30.71 | C46H46O18N4 | 963.4437 | 961.4240 | 0.87 | 945.4429; 621.3273; 455.2382; 384.1644; 293.1882; 222.1121; 165.0557 | Ddihydrocaffeoyl spermine derivative | ✓ | |||
| B74 | 31.79 | C30H45O8N3 | 576.3314 | 574.3247 | 1.45 | 544.3050; 512.2786; 412.2842; 293.1877; 222.1139; 165.0558 | Ddihydrocaffeoyl spermidine derivative | ✓ | |||
| C1 | 8.30 | C13H18O3N2 | 251.1409 | 249.1227 | 1.41 | 234.0990; 163.0697; 144.0616; 126.0558; 115.0591 | N-caffeoylputrescine isomer | [36] | ✓ | ✓ | ✓ |
| C2 | 10.26 | C13H18O3N2 | 251.1410 | 1.69 | 234.1142; 163.0402; 145.0296; 115.0876 | N-caffeoylputrescine isomer | [36] | ✓ | ✓ | ||
| C3 | 16.49 | C15H22O4N2 | 295.1674 | 1.20 | 278.1412; 207.0670; 175.0409; 147.0453; 129.1394 | coumaroyl amide derivative | [37] | ✓ | |||
| C4 | 23.48 | C15H22O4N2 | 295.1673 | 1.17 | 278.1405; 222.1142; 207.0666; 175.0412; 147.0451 | coumaroyl amide derivative | [37] | ✓ | |||
| C5 | 30.22 | C21H29O9N | 476.1905 | 474.2481 | 2.14 | 314.1381; 222.1119; 177.0545; 145.0284; 121.0648 | N-feruloyl-3-O-glucopyranosyl-tyramine ismoer | [38] | ✓ | ||
| C6 | 30.42 | C32H45O11N3 | 648.3118 | 646.2941 | 1.32 | 486.2590; 310.2119; 234.1121; 177.0544; 145.0291 | feruloyl-tyramine derivatives | ✓ | |||
| C7 | 31.30 | C17H18O4N | 302.1412 | 300.1221 | 1.46 | 286.0265; 245.8081; 165.0556; 138.0924; 121.0657 | A4 (caffeoyl-tyramine derivatives) | [32] | ✓ | ||
| C8 | 31.34 | C14H14O5N2 | 486.2587 | 484.2422 | 1.07 | 469.2316; 310.2118; 234.1120; 177.0544; 145.0283; 121.0649 | feruloyl-tyramine derivatives | ✓ | |||
| C9 | 31.83 | C21H29O9N | 476.1905 | 474.2481 | 2.07 | 314.1381; 222.1118; 177.0545; 145.0284; 121.0648 | N-feruloyl-4-O-glucopyranosyl-tyramine | [38] | ✓ | ||
| C10 | 32.09 | C36H36O8N2 | 625.2537 | 623.1578 | 1.73 | 462.1902; 351.0856; 293.08033; 201.0544; 175.0402; 149.0609; 121.0648 | Lyciumamide A | [39] | ✓ | ||
| C11 | 32.22 | C36H49O16N3 | 498.2592 | 498.2576 | 1.41 | 480.2489; 322.2119; 234.1126; 177.0545; 145.0284 | feruloyl-tyramine derivatives | ✓ | |||
| C12 | 32.57 | C17H19O4N | 300.1256 | 298.1066 | 0.48 | 253.8823; 163.0402; 121.0658 | A5 (caffeoyl-tyramine derivatives) | [32] | ✓ | ||
| C13 | 32.98 | C17H17O3N | 284.1275 | 282.1116 | 2.29 | 261.0436; 164.0705; 147.0439; 121.0648 | N-p-cis-Coumaroyl tyramine | [40] | ✓ | ✓ | |
| C14 | 33.33 | C18H19O4N | 314.1382 | 312.1221 | 1.67 | 274.8365; 243.1029; 220.0979; 195.0847; 177.0551; 145.0289; 121.0653 | N-cis-feruloyl-tyramine | [8] | ✓ | ✓ | |
| C15 | 33.47 | C17H17O3N | 284.1275 | 282.1116 | 2.29 | 261.0436; 164.0705; 147.0439; 121.0648 | N-p-trans-Coumaroyl tyramine * | ✓ | ✓ | ||
| C16 | 33.62 | C28H31O8N | 510.2116 | 508.1944 | 1.36 | 462.1963; 325.1065; 210.0545; 177.0546; 121.0648 | canabisine-H | [36] | ✓ | ||
| C17 | 33.81 | C18H19O4N | 314.1382 | 312.1221 | 1.51 | 244.0981; 220.0979; 194.0822; 177.0555; 145.0292; 121.0653 | N-trans-feruloyl-tyramine | [8] | ✓ | ✓ | ✓ |
| C18 | 34.02 | C19H19O5N | 344.1520 | 342.1323 | 1.25 | 282.3545; 177.0559; 145.0295 | A12 (feruloyl-tyramine derivatives) | [32] | ✓ | ||
| C19 | 34.98 | C36H37O9N2 | 643.2638 | 641.2465 | 1.89 | 462.1903; 325.1063; 201.0544; 177.0545; 121.0648 | feruloyl-tyramine derivatives | ✓ | |||
| C20 | 35.07 | C28H29O7N | 492.2010 | 490.1842 | 1.46 | 462.1909; 325.1066; 293.0805; 201.0546; 175.0769; 121.0649 | Lyciumamide C | [39] | ✓ | ||
| C21 | 36.57 | C54H53O12N3 | 936.3691 | 1.20 | 771.2914; 634.20612; 471.1429; 375.0859; 263.0896; 203.5736; 121.0659 | melongenamide D isomer | [41] | ✓ | |||
| D1 | 24.74 | C33H40O21 | 773.2195 | 771.1937 | 1.21 | 611.140; 464.0762; 303.0325; 163.0399; | Quercetin-3-O-Glu-7-O-Rha isomer | [42] | ✓ | ||
| D2 | 26.89 | C27H30O17 | 627.1610 | 625.1376 | 1.66 | 585.6912; 303.0520; 285.0409; 257.043; 243.5646; 201.4349; 129.02379 | Quercetin-3,7-O-diGlu | [42] | ✓ | ✓ | |
| D3 | 26.98 | C33H40O21 | 773.2195 | 771.1940 | 1.84 | 726.3508; 559.7092; 465.1061; 303.0521; 228.4964; 129.0548 | Quercetin-3-O-Soph-7-O-Rha | [42] | ✓ | ||
| D4 | 27.74 | C33H40O21 | 773.2127 | 771.1940 | 1.08 | 611.3322; 472.2473; 303.0492; 220.0965; 163.0399; 129.0541 | Quercetin-3-O-Rut-7-O-Glu | [42] | ✓ | ✓ | |
| D5 | 31.39 | C27H30O16 | 611.1600 | 609.1417 | 1.03 | 449.1113; 465.1061; 303.0492; 285.0413; 257.046; 201.0561129.05449 | Rutin * | ✓ | ✓ | ||
| D6 | 31.98 | C27H30O15 | 595.1703 | 593.1472 | 1.63 | 465.5521; 329.0679; 287.0529; 258.2196; 243.5895; 230.3383; 129.0553 | Kaempferol-3-O-Glu-7-O-Rha | [42] | ✓ | ✓ | |
| E1 | 33.67 | C45H72O17 | 885.4835 | 0.87 | 867.4724; 415.3229; 299.2362; 271.2052; 253.1947; 215.1792; 157.1011 | Gracillin | [43] | ✓ | |||
| E2 | 33.72 | C45H73O16N | 884.5084 | 882.4800 | 1.13 | 866.4896; 720.4325; 576.3898; 414.3403; 396.3259; 271.2054; 253.1949 | Solasonine * | ✓ | |||
| E3 | 33.77 | C51H84O22 | 1049.5518 | 0.90 | 887.4990; 743.3851; 417.3355; 273.2207; 255.2107 | parillin | [44] | ✓ | |||
| E4 | 33.87 | C45H75O16N | 886.5152 | 884.4957 | 0.75 | 868.5034; 722.4475; 416.356; 398.3409; 273.2205; 255.2102; 173.1323 | 5,6-dihydrosolasonine * | ✓ | |||
| E5 | 33.92 | C45H73O18 | 903.5006 | 1.21 | 741.4469; 597.3309; 417.3389; 273.2230; 255.2124; 145.0506 | Timosaponin BIII | [45] | ✓ | |||
| E6 | 34.49 | C47H77O17N | 928.5253 | 1.25 | 458.3620; 273.2207; 255.2102; 161.1323 | Lycioside B | [1] | ✓ | |||
| F1 | 3.93 | C9H11O2N | 166.0867 | 1.10 | 153.0416; 142.9681; 120.0814; 103.0548 | Phenylalanine isomer | [1] | ✓ | ✓ | ✓ | |
| F2 | 5.51 | C9H11O3N | 166.0877 | 1.13 | 142.9681; 138.0554; 120.0812; 103.0548 | Phenylalanine isomer | ✓ | ✓ | ✓ | ||
| F3 | 9.95 | C11H9O2N | 188.0721 | 1.95 | 170.0613; 146.0412; 118.0611 | 3-amino-2-naphthoic acid | [46] | ✓ | ✓ | ✓ | |
| F4 | 10.44 | 371.0991 | 0.11 | 205.0506; 163.0398; 145.9288; 119.0498 | ✓ | ||||||
| F5 | 31.16 | 568.3110 | 539.2707; 363.2378; 268.0598; 135.0436 | ✓ | |||||||
| F6 | 31.29 | 748.3361 | 609.3981; 399.1691; 360.1675; 314.1518; 215.0829; 171.0929; 136.0770 | ✓ | |||||||
| F7 | 31.69 | 630.3469 | 498.3042; 469.2647; 387.2245; 241.0723; 151.2698 | ✓ | |||||||
| F8 | 32.56 | 298.1066 | 256.0959; 178.0492; 135.0435 | ✓ | |||||||
| F9 | 32.91 | 647.3257 | 618.1393; 483.2572; 412.2080; 395.2055; 161.4154 | ✓ | |||||||
| F10 | 34.22 | 897.3956 | 895.3695 | 879.3847; 689.3096; 486.2017; 468.1911; 422.583; 395.1742; 159.0929 | ✓ | ||||||
| F11 | 34.23 | 898.4049 | 690.3203; 527.2580; 387.1645; 203.0810; 153.0655 | ✓ |
| Compounds | Fruits (μg/g) | Leaves (μg/g) | Root Barks (μg/g) |
|---|---|---|---|
| Kukoamine B | - | - | 10,900 ± 3 |
| Scopolin | 12.7 ± 0.08 | - | - |
| Chlorogenic acid | - | 1577 ± 4 | - |
| Rutin | 93 ± 5 | 663 ± 15 | - |
| Solasonine | 2.16 ± 0.02 | - | - |
| 5,6-Dihydrosolasonine | 43 ± 3 | - | - |
| N-p-trans-Coumaroyltyramine | 3.33 ± 0.02 | 14 ± 1 | - |
| Extracts | DPPH(IC50) 1 (µg/mL) | ABTS(IC50) 2 (µg/mL) | FRAP(RC50) 3 (µg/mL) |
|---|---|---|---|
| Fruits | 1974.3 ± 0.4 | 247.0 ± 0.8 | 725 ± 1.4 |
| Leaves | 123.5 ± 0.5 | 56 ± 1 | 192.6 ± 0.02 |
| Root barks | 85.0 ± 0.3 | 40.7 ± 0.4 | 224 ± 1 |
| Ascorbic acid 4 | 11.2 ± 0.5 | - | - |
| Trolox 5 | - | 6 ± 1 | 37.7 ± 0.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Ren, W.; Zhang, N.; Bing, T.; Liu, X.; Zhao, Z.; Shangguan, D. Comparative Study of the Chemical Constituents and Bioactivities of the Extracts from Fruits, Leaves and Root Barks of Lycium barbarum. Molecules 2019, 24, 1585. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24081585
Xiao X, Ren W, Zhang N, Bing T, Liu X, Zhao Z, Shangguan D. Comparative Study of the Chemical Constituents and Bioactivities of the Extracts from Fruits, Leaves and Root Barks of Lycium barbarum. Molecules. 2019; 24(8):1585. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24081585
Chicago/Turabian StyleXiao, Xiao, Wei Ren, Nan Zhang, Tao Bing, Xiangjun Liu, Zhenwen Zhao, and Dihua Shangguan. 2019. "Comparative Study of the Chemical Constituents and Bioactivities of the Extracts from Fruits, Leaves and Root Barks of Lycium barbarum" Molecules 24, no. 8: 1585. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24081585