Checking NEKs: Overcoming a Bottleneck in Human Diseases
Abstract
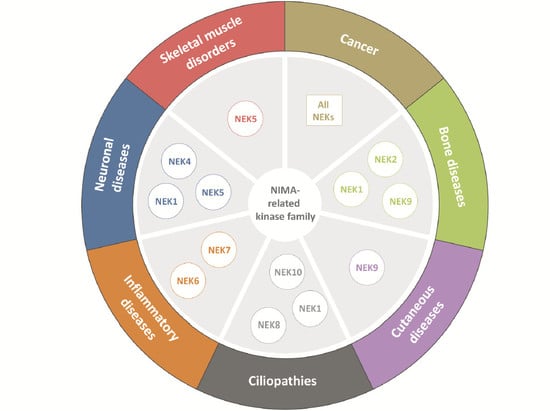
:1. Introduction
2. NEK1
3. NEK2
4. NEK3
5. NEK4
6. NEK5
7. NEK6
8. NEK7
9. NEK8
10. NEK9
11. NEK10
12. NEK11
13. Pharmacological Inhibitors of the NEK Family
14. Conclusions
Funding
Conflicts of Interest
References
- Oakley, B.R.; Morris, N.R. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 1983, 96, 1155–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osmani, S.A.; May, G.S.; Morris, N.R. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 1987, 104, 1495–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.P.; Kemp, B.E.; Means, A.R. Identification of substrate specificity determinants for the cell cycle- regulated NIMA protein kinase. J. Biol. Chem. 1994, 269, 6603–6607. [Google Scholar] [PubMed]
- Moniz, L.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 18, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Osmani, S.A.; Mirabito, P.M. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 1998, 141, 1575–1587. [Google Scholar] [CrossRef] [Green Version]
- Salaun, P.; Rannou, Y.; Claude, P. Cdk1, plks, auroras, and neks: The mitotic bodyguards. Adv. Exp. Med. Biol. 2008, 617, 41–56. [Google Scholar]
- Meirelles, G.V.; Perez, A.M.; de Souza, E.E.; Basei, F.L.; Papa, P.F.; Melo Hanchuk, T.D.; Cardoso, V.B.; Kobarg, J. “Stop Ne(c)king around”: How interactomics contributes to functionally characterize Nek family kinases. World J. Biol. Chem. 2014, 5, 141–160. [Google Scholar]
- Thiel, C.; Kessler, K.; Giessl, A.; Dimmler, A.; Shalev, S.A.; von der Haar, S.; Zenker, M.; Zahnleiter, D.; Stöss, H.; Beinder, E.; et al. NEK1 mutations cause short-rib polydactyly syndrome type majewski. Am. J. Hum. Genet. 2011, 88, 106–114. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; Schultz, S.J.; Bartek, J.; Nigg, E.A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 1995, 270, 12899–12905. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Nigg, E.A. Cloning and characterization of the murine Nek3 protein kinase, a novel member of the NIMA family of putative cell cycle regulators. J. Biol. Chem. 1999, 274, 13491–13497. [Google Scholar] [CrossRef] [Green Version]
- O’Regan, L.; Fry, A.M. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 2009, 29, 3975–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, E.E.; Meirelles, G.V.; Godoy, B.B.; Perez, A.M.; Smetana, J.H.C.; Doxsey, S.J.; McComb, M.E.; Costello, C.E.; Whelan, S.A.; Kobarg, J. Characterization of the human NEK7 interactome suggests catalytic and regulatory properties distinct from those of NEK6. J. Proteome Res. 2014, 13, 4074–4090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moniz, L.S.; Stambolic, V. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol. Cell. Biol. 2011, 31, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belham, C.; Roig, J.; Caldwell, J.A.; Aoyama, Y.; Kemp, B.E.; Comb, M.; Avruch, J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 2003, 278, 34897–34909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, E.E.; Hehnly, H.; Perez, A.M.; Meirelles, G.V.; Smetana, J.H.C.; Doxsey, S.; Kobarg, J. Human Nek7-interactor RGS2 is required for mitotic spindle organization Human Nek7-interactor RGS2 is required for mitotic spindle organization. Cell Cycle 2015, 14, 656–667. [Google Scholar] [CrossRef]
- Roig, J.; Groen, A.; Caldwell, J.; Avruch, J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell 2005, 16, 4827–4840. [Google Scholar] [CrossRef] [Green Version]
- Letwin, K.; Mizzen, L.; Motro, B.; Ben-David, Y.; Bernstein, A.; Pawson, T. A mammalian dual specificity protein kinase, Nek1, is related to the NIMA cell cycle regulator and highly expressed in meiotic germ cells. EMBO J. 1992, 11, 3521–3531. [Google Scholar] [CrossRef]
- Basei, F.L.; Meirelles, G.V.; Righetto, G.L.; Dos Santos Migueleti, D.L.; Smetana, J.H.C.; Kobarg, J. New interaction partners for Nek4.1 and Nek4.2 isoforms: From the DNA damage response to RNA splicing. Proteome Sci. 2015, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Wang, Z.-B.; Wang, H.-H.; Zhang, T.; Qi, S.-T.; Ouyang, Y.-C.; Hou, Y.; Sun, Q.-Y. Nek11 regulates asymmetric cell division during mouse oocyte meiotic maturation. Biochem. Biophys. Res. Commun. 2016, 474, 667–672. [Google Scholar] [CrossRef]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Prosser, S.L.; Sahota, N.K.; Pelletier, L.; Morrison, C.G.; Fry, A.M. Nek5 promotes centrosome integrity in interphase and loss of centrosome cohesion in mitosis. J. Cell Biol. 2015, 209, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Holloway, K.; Roberson, E.C.; Corbett, K.L.; Kolas, N.K.; Nieves, E.; Cohen, P.E. NEK1 facilitates cohesin removal during mammalian spermatogenesis. Genes (Basel) 2011, 2, 260–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhya, P.; Birkenmeier, E.H.; Birkenmeier, C.S.; Barker, J.E. Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. USA 2000, 97, 217–221. [Google Scholar] [CrossRef] [Green Version]
- Joseph Endicott, S.; Basu, B.; Khokha, M.; Brueckner, M. The nima-like kinase nek2 is a key switch balancing cilia biogenesis and resorption in the development of left-right asymmetry. Devlopment 2015, 142, 4068–4079. [Google Scholar]
- Doles, J.; Hemann, M.T. NEK4 status influences differential sensitivity to microtubule poisons. Cancer Res. 2011, 70, 1033–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porpora, M.; Sauchella, S.; Rinaldi, L.; Delle Donne, R.; Sepe, M.; Torres-Quesada, O.; Intartaglia, D.; Garbi, C.; Insabato, L.; Santoriello, M.; et al. Counterregulation of cAMP-directed kinase activities controls ciliogenesis. Nat. Commun. 2018, 1224, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, K.; Sawasaki, T. Nek5, a novel substrate for caspase-3, promotes skeletal muscle differentiation by up-regulating caspase activity. FEBS Lett. 2013, 587, 2219–2225. [Google Scholar] [CrossRef] [Green Version]
- Gerçeker, E.; Boyacioglu, S.O.; Kasap, E.; Baykan, A.; Yuceyar, H.; Yildirim, H.; Ayhan, S.; Ellidokuz, E.; Korkmaz, M. Never in mitosis gene A-related kinase 6 and aurora kinase A: New gene biomarkers in the conversion from ulcerative colitis to colorectal cancer. Oncol. Rep. 2015, 34, 1905–1914. [Google Scholar] [CrossRef]
- Gross, O.; Thomas, C.J.; Guarda, G.; Tschopp, J. The inflammasome: An integrated view. Immunol. Rev. 2011, 243, 136–151. [Google Scholar] [CrossRef]
- Melo-Hanchuk, T.D.; Slepicka, P.F.; Meirelles, G.V.; Basei, F.L.; Lovato, D.V.; Granato, D.C.; Pauletti, B.A.; Domingues, R.R.; Leme, A.F.P.; Pelegrini, A.L.; et al. NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin. Sci. Rep. 2017, 7, 5445. [Google Scholar] [CrossRef]
- Nguyen, C.L.; Possemato, R.; Bauerlein, E.L.; Xie, A.; Scully, R.; Hahn, W.C. Nek4 regulates entry into replicative senescence and the response to DNA damage in human fibroblasts. Mol. Cell. Biol. 2012, 32, 3963–3977. [Google Scholar] [CrossRef] [Green Version]
- Melo-Hanchuk, T.D.; Slepicka, P.F.; Pelegrini, A.L.; Menck, C.F.M.; Kobarg, J. NEK5 interacts with topoisomerase IIβ and is involved in the DNA damage response induced by etoposide. J. Cell. Biochem. 2019, 120, 16853–16866. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.C.; Lin, J.R.; Vannier, J.B.; Slaats, G.G.; Kile, A.C.; Paulsen, R.D.; Manning, D.K.; Beier, D.R.; Giles, R.H.; Boulton, S.J.; et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell 2013, 51, 423–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noguchi, K.; Fukazawa, H.; Murakami, Y.; Uehara, Y. Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses. J. Biol. Chem. 2002, 277, 39655–39665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malumbres, M. Physiological relevance of cell cycle kinases. Physiol. Rev. 2011, 91, 973–1007. [Google Scholar] [CrossRef]
- de Carcer, G.; Perez de Castro, I.; Malumbres, M. Targeting Cell Cycle Kinases for Cancer Therapy. Curr. Med. Chem. 2007, 14, 969–985. [Google Scholar] [CrossRef]
- O’Regan, L.; Blot, J.; Fry, A.M. Mitotic regulation by NIMA-related kinases. Cell Div. 2007, 2, 25. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef] [Green Version]
- Wells, C.I.; Kapadia, N.R.; Couñago, R.M.; Drewry, D.H. In depth analysis of kinase cross screening data to identify chemical starting points for inhibition of the Nek family of kinases. Medchemcomm 2018, 9, 44–66. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Lasseigne, B.N.; Petrovski, S.; Sapp, P.C.; Dion, P.A.; Leblond, C.S.; Couthouis, J.; Lu, Y.F.; Wang, Q.; Krueger, B.J.; et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science (80-.) 2015, 347, 1436–1441. [Google Scholar] [CrossRef] [Green Version]
- Kenna, K.P.; van Doormaal, P.T.C.; Dekker, A.M.; Ticozzi, N.; Kenna, B.J.; Diekstra, F.P.; van Rheenen, W.; van Eijk, K.R.; Jones, A.R.; Keagle, P.; et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 2016, 48, 1037–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, D.; Müller, K.; Wieland, T.; Weydt, P.; Böhm, S.; Lulé, D.; Hübers, A.; Neuwirth, C.; Weber, M.; Borck, G.; et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain 2016, 136, e28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gratten, J.; Zhao, Q.; Benyamin, B.; Garton, F.; He, J.; Leo, P.J.; Mangelsdorf, M.; Anderson, L.; Zhang, Z.-H.; Chen, L.; et al. Whole-exome sequencing in amyotrophic lateral sclerosis suggests NEK1 is a risk gene in Chinese. Genome Med. 2017, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Lei, X.; Liu, F.; Cui, B.; Liu, Q.; Ding, Q.; Liu, M.S.; Li, X.G.; Cui, L.; Zhang, X. Mutation screening of NEK1 in Chinese ALS patients. Neurobiol. Aging 2018, 71, 267.e1–267.e4. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Van Mossevelde, S.; Dillen, L.; De Bleecker, J.L.; Moisse, M.; Van Damme, P.; Van Broeckhoven, C.; van der Zee, J. BELNEU Consortium NEK1 genetic variability in a Belgian cohort of ALS and ALS-FTD patients. Neurobiol. Aging 2018, 61, 255.e1–255.e7. [Google Scholar] [CrossRef]
- Higelin, J.; Catanese, A.; Semelink-Sedlacek, L.L.; Oeztuerk, S.; Lutz, A.-K.; Bausinger, J.; Barbi, G.; Speit, G.; Andersen, P.M.; Ludolph, A.C.; et al. NEK1 loss-of-function mutation induces DNA damage accumulation in ALS patient-derived motoneurons. Stem Cell Res. 2018, 30, 150–162. [Google Scholar] [CrossRef]
- Surpili, M.J.; Delben, T.M.; Kobarg, J. Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry 2003, 42, 15369–15376. [Google Scholar] [CrossRef]
- Liu, S.; Ho, C.K.; Ouyang, J.; Zou, L. Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2175–2180. [Google Scholar] [CrossRef] [Green Version]
- Spies, J.; Waizenegger, A.; Barton, O.; Sürder, M.; Wright, W.D.; Heyer, W.D.; Löbrich, M. Nek1 Regulates Rad54 to Orchestrate Homologous Recombination and Replication Fork Stability. Mol. Cell 2016, 62, 903–917. [Google Scholar] [CrossRef] [Green Version]
- Waters, A.M.; Beales, P.L. Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 2011, 26, 1039–1056. [Google Scholar] [CrossRef] [Green Version]
- Valente, E.M.; Rosti, R.O.; Gibbs, E.; Gleeson, J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014, 10, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McInerney-Leo, A.M.; Harris, J.E.; Leo, P.J.; Marshall, M.S.; Gardiner, B.; Kinning, E.; Leong, H.Y.; McKenzie, F.; Ong, W.P.; Vodopiutz, J.; et al. Whole exome sequencing is an efficient, sensitive and specific method for determining the genetic cause of short-rib thoracic dystrophies. Clin. Genet. 2015, 88, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Monroe, G.R.; Kappen, I.F.P.M.; Stokman, M.F.; Terhal, P.A.; Van Den Boogaard, M.H.; Savelberg, S.M.C.; Van Der Veken, L.T.; Van Es, R.J.J.; Lens, S.M.; Hengeveld, R.C.; et al. Compound heterozygous NEK1 variants in two siblings with oral-facial-digital syndrome type II (Mohr syndrome). Eur. J. Hum. Genet. 2016, 24, 1752–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-P.; Ko, T.-M.; Chang, T.-Y.; Chern, S.-R.; Chen, S.-W.; Lai, S.-T.; Chuang, T.-Y.; Wang, W. Prenatal diagnosis of short-rib polydactyly syndrome type III or short-rib thoracic dysplasia 3 with or without polydactyly (SRTD3) associated with compound heterozygous mutations in DYNC2H1 in a fetus. Taiwan. J. Obstet. Gynecol. 2018, 57, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-P.; Chern, S.-R.; Chang, T.-Y.; Su, Y.-N.; Chen, Y.-Y.; Su, J.-W.; Wang, W. Prenatal diagnosis and molecular genetic analysis of short rib-polydactyly syndrome type III (Verma-Naumoff) in a second-trimester fetus with a homozygous splice site mutation in intron 4 in the NEK1 gene. Taiwan. J. Obstet. Gynecol. 2012, 51, 266–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, T. Understanding high grade glioma: Molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013, 331, 139–146. [Google Scholar] [CrossRef]
- Zhu, J.; Cai, Y.; Liu, P.; Zhao, W. Frequent Nek1 overexpression in human gliomas. Biochem. Biophys. Res. Commun. 2016, 476, 522–527. [Google Scholar] [CrossRef]
- Cabral de Almeida Cardoso, L.; Rodriguez-Laguna, L.; Del Carmen Crespo, M.; Vallespín, E.; Palomares-Bralo, M.; Martin-Arenas, R.; Rueda-Arenas, I.; Silvestre de Faria, P.A.; GT-CSGP Working Group; García-Miguel, P.; et al. Array CGH Analysis of Paired Blood and Tumor Samples from Patients with Sporadic Wilms Tumor. PLoS ONE 2015, 10, e0136812. [Google Scholar] [CrossRef] [PubMed]
- Cardote, T.A.F.; Gadd, M.S.; Ciulli, A. Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex. Structure 2017, 25, 901–911.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, M.; Pabla, N.; Huang, S.; Dong, Z. Nek1 phosphorylates Von Hippel-Lindau tumor suppressor to promote its proteasomal degradation and ciliary destabilization. Cell Cycle 2013, 12, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Zhou, J.; Chen, J.; Zhu, J.; Liu, S.C.; Ding, X.F.; Zhang, Q. VHL regulates NEK1 via both HIF-2α pathway and ubiquitin-proteasome pathway in renal cancer cell. Biochem. Biophys. Res. Commun. 2019, 509, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett. 2019, 453, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. Targeting the TLK1/NEK1 DDR axis with Thioridazine suppresses outgrowth of androgen independent prostate tumors. Int. J. Cancer 2019, 145, 1055–1067. [Google Scholar] [CrossRef] [Green Version]
- Melo-Hanchuk, T.D.; Martins, M.B.; Cunha, L.L.; Soares, F.A.; Ward, L.S.; Vassallo, J.; Kobarg, J. Expression of the NEK family in normal and cancer tissue: An immunohistochemical study. BMC Cancer 2020, 20, 23. [Google Scholar] [CrossRef]
- Gönczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639–652. [Google Scholar] [CrossRef]
- Hames, R.S.; Fry, A.M. Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochem. J. 2002, 361, 77–85. [Google Scholar] [CrossRef]
- Ha Kim, Y.; Yeol Choi, J.; Jeong, Y.; Wolgemuth, D.J.; Rhee, K. Nek2 localizes to multiple sites in mitotic cells, suggesting its involvement in multiple cellular functions during the cell cycle. Biochem. Biophys. Res. Commun. 2002, 290, 730–736. [Google Scholar] [CrossRef]
- Dasgupta, A.; Amack, J.D. Cilia in vertebrate left - Right patterning. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 1710. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, R.; Oborski, M.J.; Hwang, M.; Lieberman, F.S.; Mountz, J.M. Malignant gliomas: Current perspectives in diagnosis, treatment, and early response assessment using advanced quantitative imaging methods. Cancer Manag. Res. 2014, 6, 149–170. [Google Scholar] [PubMed] [Green Version]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA J. Am. Med. Assoc. 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, B.; Hou, X.; Pang, B.; Guo, P.; Jiang, W.; Ding, Q.; Zhang, R.; Xin, T.; Guo, H.; et al. Overexpression of NIMA-related kinase 2 is associated with poor prognoses in malignant glioma. J. Neurooncol. 2017, 132, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, P.; Pavlyukov, M.S.; Yu, H.; Zhang, Z.; Kim, S.-H.; Minata, M.; Mohyeldin, A.; Xie, W.; Chen, D.; et al. Targeting NEK2 attenuates glioblastoma growth and radioresistance by destabilizing histone methyltransferase EZH2. J. Clin. Investig. 2017, 127, 3075–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pacific J. Cancer Prev. 2017, 18, 3–9. [Google Scholar]
- Orzan, F.; Pellegatta, S.; Poliani, P.L.; Pisati, F.; Caldera, V.; Menghi, F.; Kapetis, D.; Marras, C.; Schiffer, D.; Finocchiaro, G. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol. Appl. Neurobiol. 2011, 37, 381–394. [Google Scholar] [CrossRef]
- Smith, L.; McCourt, O.; Henrich, M.; Paton, B.; Yong, K.; Wardle, J.; Fisher, A. Multiple myeloma and physical activity: A scoping review. BMJ Open 2015, 5, e009576. [Google Scholar] [CrossRef] [Green Version]
- Nikesitch, N.; Ling, S.C.W. Molecular mechanisms in multiple myeloma drug resistance. J. Clin. Pathol. 2016, 69, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Franqui-Machin, R.; Hao, M.; Bai, H.; Gu, Z.; Zhan, X.; Habelhah, H.; Jethava, Y.; Qiu, L.; Frech, I.; Tricot, G.; et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Investig. 2018, 128, 2877–2893. [Google Scholar] [CrossRef]
- Hao, M.; Franqui-Machin, R.; Xu, H.; Shaughnessy, J.; Barlogie, B.; Roodman, D.; Quelle, D.E.; Janz, S.; Tomasson, M.H.; Sanderson, R.D.; et al. NEK2 induces osteoclast differentiation and bone destruction via heparanase in multiple myeloma. Leukemia 2017, 31, 1648–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Miao, Z.; Yang, Q.; Wang, Y.; Zhang, J. The Dynamic Roles of Mesenchymal Stem Cells in Colon Cancer. Can. J. Gastroenterol. Hepatol. 2018, 2018, 7628763. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhai, X.; Yuan, R. Clinical significance and prognostic value of Nek2 protein expression in colon cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 15467–15473. [Google Scholar] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [Green Version]
- Balogh, J.; Victor, D., III; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P., Jr. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Lai, X.-B.; Nie, Y.-Q.; Huang, H.-L.; Li, Y.-F.; Cao, C.-Y.; Yang, H.; Shen, B.; Feng, Z.-Q. NIMA-related kinase 2 regulates hepatocellular carcinoma cell growth and proliferation. Oncol. Lett. 2017, 13, 1587–1594. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.-J.; Chen, J.; Ji, F.; Ju, W.-Q.; Zhao, Q.; Chen, M.-G.; Guo, Z.-Y.; Wu, L.-W.; Ma, Y.; Wang, D.-P.; et al. MiR-486-5p negatively regulates oncogenic NEK2 in hepatocellular carcinoma. Oncotarget 2017, 8, 52948–52959. [Google Scholar] [CrossRef] [Green Version]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45. [Google Scholar] [CrossRef]
- Lin, S.; Zhou, S.; Jiang, S.; Liu, X.; Wang, Y.; Zheng, X.; Zhou, H.; Li, X.; Cai, X. NEK2 regulates stem-like properties and predicts poor prognosis in hepatocellular carcinoma. Oncol. Rep. 2016, 36, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.; Liu, Y.; Yang, M.; Yang, K.; Huang, J.; Feng, D. Increased NEK2 in hepatocellular carcinoma promotes cancer progression and drug resistance by promoting PP1/Akt and Wnt activation. Oncol. Rep. 2016, 36, 2193–2199. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhong, Y.; Shen, Q.; Zhou, Y.; Deng, X.; Li, C.; Chen, J.; Zhou, Y.; He, M. NEK2 serves as a prognostic biomarker for hepatocellular carcinoma. Int. J. Oncol. 2017, 50, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, W.; Wang, Y.; Huang, X.; Zhang, Z.; Chen, B.; Xie, W.; Li, S.; Shen, S.; Peng, B. NEK2 promotes hepatocellular carcinoma migration and invasion through modulation of the epithelial-mesenchymal transition. Oncol. Rep. 2018, 39, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-M.; Lin, S.-L.; Lee, K.-Y.; Chuang, H.-C.; Feng, P.-H.; Cheng, W.-L.; Liao, C.-J.; Chi, H.-C.; Lin, Y.-H.; Tsai, C.-Y.; et al. Hepatoma cell functions modulated by NEK2 are associated with liver cancer progression. Int. J. Cancer 2017, 140, 1581–1596. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Fisher, D.E.; Garbe, C.; Gershenwald, J.E.; Grob, J.-J.; Halpern, A.; Herlyn, M.; Marchetti, M.A.; McArthur, G.; Ribas, A.; et al. Melanoma. Nat. Rev. Dis. Prim. 2015, 1, 15003. [Google Scholar] [CrossRef]
- Owens, B. Melanoma. Nature 2014, 515, S109. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Sun, S.-G.; Hou, S. Aberrant NEK2 expression might be an independent predictor for poor recurrence-free survival and overall survival of skin cutaneous melanoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3694–3702. [Google Scholar]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet (Lond. Engl.) 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Kokuryo, T.; Hibino, S.; Suzuki, K.; Watanabe, K.; Yokoyama, Y.; Nagino, M.; Senga, T.; Hamaguchi, M. Nek2 siRNA therapy using a portal venous port–catheter system for liver metastasis in pancreatic cancer. Cancer Sci. 2016, 107, 1315–1320. [Google Scholar] [CrossRef] [Green Version]
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal carcinoma. Lancet (Lond. Engl.) 2016, 387, 1012–1024. [Google Scholar] [CrossRef]
- Petersson, F. Nasopharyngeal carcinoma: A review. Semin. Diagn. Pathol. 2015, 32, 54–73. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zeng, L.; Guan, Y.; Feng, X.; Zhu, Y.; Lu, Y.; Shi, C.; Chen, S.; Xia, J.; Guo, J.; et al. High NEK2 confers to poor prognosis and contributes to cisplatin-based chemotherapy resistance in nasopharyngeal carcinoma. J. Cell. Biochem. 2019, 120, 3547–3558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.; Paz-ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Nanavaty, P.; Alvarez, M.S.; Alberts, W.M. Lung cancer screening: Advantages, controversies, and applications. Cancer Control 2014, 21, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.-X.; Yin, J.-Y.; Shen, Y.; Zhang, W.; Zhou, H.-H.; Liu, Z.-Q. Genome-scale analysis identifies NEK2, DLGAP5 and ECT2 as promising diagnostic and prognostic biomarkers in human lung cancer. Sci. Rep. 2017, 7, 8072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayward, D.G.; Clarke, R.B.; Faragher, A.J.; Pillai, M.R.; Hagan, I.M.; Fry, A.M. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004, 64, 7370–7376. [Google Scholar] [CrossRef] [Green Version]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet (Lond. Engl.) 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018: Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Fukasawa, K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005, 230, 6–19. [Google Scholar] [CrossRef]
- Chan, J.Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 2011, 7, 1122–1144. [Google Scholar] [CrossRef]
- Fry, A.M. The Nek2 protein kinase: A novel regulator of centrosome structure. Oncogene 2002, 6184–6194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marina, M.; Saavedra, H.I. Nek2 and Plk4: Prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. Landmark 2014, 19, 352–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison Pitner, M.K.; Saavedra, H.I. Cdk4 and Nek2 Signal Binucleation and Centrosome Amplification in a Her2+ Breast Cancer Model. PLoS ONE 2013, 8, e65971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, L.; Van Steensel, B.; Broccoli, D.; Erdjument-Bromage, H.; Hanish, J.; Tempst, P.; De Lange, T. A human telomeric protein. Science (80-.) 1995, 270, 1663–1667. [Google Scholar] [CrossRef]
- Shen, M.; Haggblom, C.; Vogt, M.; Hunter, T.; Lu, K.P. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc. Natl. Acad. Sci. USA 1997, 94, 13618–13623. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Gollahon, L. Mitotic perturbations induced by Nek2 overexpression require interaction with TRF1 in breast cancer cells. Cell Cycle 2013, 12, 3599–3614. [Google Scholar] [CrossRef] [Green Version]
- Cappello, P.; Blaser, H.; Gorrini, C.; Lin, D.C.C.; Elia, A.J.; Wakeham, A.; Haider, S.; Boutros, P.C.; Mason, J.M.; Miller, N.A.; et al. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene 2014, 33, 2375–2384. [Google Scholar] [CrossRef] [Green Version]
- Nuncia-Cantarero, M.; Martinez-Canales, S.; Andrés-Pretel, F.; Santpere, G.; Ocaña, A.; Galan-Moya, E.M. Functional transcriptomic annotation and protein–protein interaction network analysis identify NEK2, BIRC5, and TOP2A as potential targets in obese patients with luminal A breast cancer. Breast Cancer Res. Treat. 2018, 168, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M.; Okano, Y. Molecular cloning and characterization of the human NIMA-related protein kinase 3 gene (NEK3). Cytogenet. Cell Genet. 2001, 95, 177–182. [Google Scholar] [CrossRef]
- SJ, S.; Andrew, M.F.; Christine, S.; T, R.; Erich, A.N. Cell cycledependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 1994, 5, 625–635. [Google Scholar]
- Chang, J.; Baloh, R.H.; Milbrandt, J. The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J. Cell Sci. 2009, 122, 2274–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.L.; DeMaria, J.E.; Freier, D.O.; Riegel, A.M.; Clevenger, C.V. Novel association of Vav2 and Nek3 modulates signaling through the human prolactin receptor. Mol. Endocrinol. 2005, 19, 939–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, V.B.; Melo-Hanchuk, T.D.; de Souza, E.E.; Papa, P.F.; Meirelles, G.V.; Kobarg, J. Identification of NEK3 interacting proteins and functional characterization of its signaling mechanisms. J. Integr. OMICS 2017, 7. [Google Scholar] [CrossRef]
- Clevenger, C.V.; Ngo, W.; Sokol, D.L.; Luger, S.M.; Gewirtz, A.M. Vav is necessary for prolactin-stimulated proliferation and is translocated into the nucleus of a T-cell line. J. Biol. Chem. 1995, 270, 13246–13253. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.L.; Antico, G.; Raghunath, P.N.; Tomaszewski, J.E.; Clevenger, C.V. Nek3 kinase regulates prolactin-mediated cytoskeletal reorganization and motility of breast cancer cells. Oncogene 2007, 26, 4668–4678. [Google Scholar] [CrossRef] [Green Version]
- Iarc-International Agency for Research on Cancer. Available online: https://www.iarc.fr/ (accessed on 28 March 2020).
- Ang, T.L.; Fock, K.M. Clinical epidemiology of gastric cancer. Singapore Med. J. 2014, 55, 621–628. [Google Scholar] [CrossRef] [Green Version]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, G.J.A.; Maciejewski, R.; Polkowski, W.P. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Song, J.; Chen, J.; Xiao, J.; Ni, J.; Wu, C. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine (Baltimore) 2018, 97, e9630. [Google Scholar] [CrossRef]
- Coene, K.L.M.; Mans, D.A.; Boldt, K.; Gloeckner, C.J.; van Reeuwijk, J.; Bolat, E.; Roosing, S.; Letteboer, S.J.F.; Peters, T.A.; Cremers, F.P.M.; et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum. Mol. Genet. 2011, 20, 3592–3605. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, R.; Szymanska, K.; Basu, B.; Patel, N.; Ewida, N.; Faqeih, E.; Al Hashem, A.; Derar, N.; Alsharif, H.; Aldahmesh, M.A.; et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016, 17, 242. [Google Scholar] [CrossRef] [Green Version]
- Huo, T.; Canepa, R.; Sura, A.; Modave, F.; Gong, Y. Colorectal cancer stages transcriptome analysis. PLoS ONE 2017, 12, e0188697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, N.H.; Zhang, L.; Xiao, Z.; Rong, Z.X.; Li, Z.; He, J.; Chen, L.; Ou, D.M.; Liao, W.H.; Sun, L.Q. NEK4 kinase regulates EMT to promote lung cancer metastasis. J. Cell. Mol. Med. 2018, 22, 5877–5887. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, A.S.; Sharova, E.I.; Danilenko, S.A.; Butusova, T.B.; Vasiliev, A.O.; Govorov, A.V.; Prilepskaya, E.A.; Pushkar, D.Y.; Kostryukova, E.S. Novel RNA biomarkers of prostate cancer revealed by RNA-seq analysis of formalin-fixed samples obtained from Russian patients. Oncotarget 2017, 8, 32990–33001. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Zhang, J.; Yang, X.; Wu, Z.; Sun, C.; Wang, Z.; Wang, B. NEK5 promotes breast cancer cell proliferation through up-regulation of Cyclin A2. Mol. Carcinog. 2019, 58, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Vaz Meirelles, G.; Ferreira Lanza, D.C.; da Silva, J.C.; Santana Bernachi, J.; Paes Leme, A.F.; Kobarg, J. Characterization of hNek6 interactome reveals an important role for its short N-terminal domain and colocalization with proteins at the centrosome. J. Proteome Res. 2010, 9, 6298–6316. [Google Scholar] [CrossRef]
- Meirelles, G.V.; Silva, J.C.; Mendonça, Y.D.A.; Ramos, C.H.I.; Torriani, I.L.; Kobarg, J. Human Nek6 is a monomeric mostly globular kinase with an unfolded short N-terminal domain. BMC Struct. Biol. 2011, 11, 12. [Google Scholar] [CrossRef] [Green Version]
- Richards, M.W.; O’Regan, L.; Mas-Droux, C.; Blot, J.M.Y.; Cheung, J.; Hoelder, S.; Fry, A.M.; Bayliss, R. An Autoinhibitory Tyrosine Motif in the Cell-Cycle-Regulated Nek7 Kinase Is Released through Binding of Nek9. Mol. Cell 2009, 36, 560–570. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, H.; Wang, D.; Han, S.; Wang, K.; Yao, A.; Li, X. Never in mitosis gene A-related kinase 6 promotes cell proliferation of hepatocellular carcinoma via cyclin B modulation. Oncol. Lett. 2014, 8, 1163–1168. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, A.D.; Schinzel, A.C.; Cotter, M.B.; Lis, R.T.; Labella, K.; Lock, Y.J.; Izzo, F.; Guney, I.; Bowden, M.; Li, Y.Y.; et al. Castration Resistance in Prostate Cancer Is Mediated by the Kinase NEK6. Cancer Res. 2017, 77, 753–765. [Google Scholar] [CrossRef] [Green Version]
- Orenay-Boyacioglu, S.; Kasap, E.; Gerceker, E.; Yuceyar, H.; Demirci, U.; Bilgic, F.; Korkmaz, M. Expression profiles of histone modification genes in gastric cancer progression. Mol. Biol. Rep. 2018, 45, 2275–2282. [Google Scholar] [CrossRef]
- Xu, J.; He, Q.; He, X.; Shao, Q.; Tao, H.; Ye, Z. Expression of NEK-6 in gastric cancer and its clinical significance. Zhonghua Wei Chang Wai Ke Za Zhi 2015, 18, 1036–1040. [Google Scholar] [PubMed]
- Takeno, A.; Takemasa, I.; Doki, Y.; Yamasaki, M.; Miyata, H.; Takiguchi, S.; Fujiwara, Y.; Matsubara, K.; Monden, M. Integrative approach for differentially overexpressed genes in gastric cancer by combining large-scale gene expression profiling and network analysis. Br. J. Cancer 2008, 99, 1307–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shussman, N.; Wexner, S.D. Colorectal polyps and polyposis syndromes. Gastroenterol. Rep. 2014, 2, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasap, E.; Gerceker, E.; Boyacıoglu, S.Ö.; Yuceyar, H.; Yıldırm, H.; Ayhan, S.; Korkmaz, M. The potential role of the NEK6, AURKA, AURKB, and PAK1 genes in adenomatous colorectal polyps and colorectal adenocarcinoma. Tumour Biol. 2016, 37, 3071–3080. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Ni, X.; Xia, L.; Shao, Z. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in Human Breast Cancer. Pathol. Res. Pract. 2018, 214, 1648–1654. [Google Scholar] [CrossRef]
- De Donato, M.; Fanelli, M.; Mariani, M.; Raspaglio, G.; Pandya, D.; He, S.; Fiedler, P.; Petrillo, M.; Scambia, G.; Ferlini, C. Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Am. J. Cancer Res. 2015, 5, 1862–1877. [Google Scholar]
- Chen, F.; Feng, Z.; Zhu, J.; Liu, P.; Yang, C.; Huang, R.; Deng, Z. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol. Ther. 2018, 19, 1139–1152. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Chen, Z.; Xing, Y. MiR-506-3p inhibits cell proliferation, induces cell cycle arrest and apoptosis in retinoblastoma by directly targeting NEK6. Cell Biol. Int. 2018. [Google Scholar] [CrossRef]
- Davidson, B.; Tropé, C.G. Ovarian cancer: Diagnostic, biological and prognostic aspects. Womens. Health (Lond. Engl.) 2014, 10, 519–533. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Salanti, G.; Pavlidis, N.; Paraskevaidis, E.; Ioannidis, J.P.A. Survival Bene fi ts With Diverse Chemotherapy Regimens for Ovarian Cancer: Meta-analysis of Multiple Treatments. J. Natl. Cancer Inst. 2006, 98, 1655–1663. [Google Scholar] [CrossRef] [Green Version]
- Rohwer, N.; Cramer, T. Hypoxia-mediated drug resistance: Novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist. Updat. 2011, 14, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, N.; Zuo, L.; Chen, H.; Zhang, C.; He, P.; Yan, H. Long non-coding RNA homeobox A11 antisense RNA (HOXA11-AS) promotes retinoblastoma progression via sponging miR-506-3p. Onco Targets Ther. 2019, 12, 3509–3517. [Google Scholar] [CrossRef] [Green Version]
- Haq, T.; Richards, M.W.; Burgess, S.G.; Gallego, P.; Yeoh, S.; O’Regan, L.; Reverter, D.; Roig, J.; Fry, A.M.; Bayliss, R. Mechanistic basis of Nek7 activation through Nek9 binding and induced dimerization. Nat. Commun. 2015, 6, 8771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandli, M.; Feige, E.; Chen, A.; Kilfin, G.; Motro, B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics 2000, 68, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Freixo, F.; Martinez Delgado, P.; Manso, Y.; Sánchez-Huertas, C.; Lacasa, C.; Soriano, E.; Roig, J.; Lüders, J. NEK7 regulates dendrite morphogenesis in neurons via Eg5-dependent microtubule stabilization. Nat. Commun. 2018, 9, 2330. [Google Scholar] [CrossRef]
- Wolfe, M.S. Tau mutations in neurodegenerative diseases. J. Biol. Chem. 2009, 284, 6021–6025. [Google Scholar] [CrossRef] [Green Version]
- Denton, K.R.; Lei, L.; Grenier, J.; Rodionov, V.; Blackstone, C.; Li, X.J. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells 2014, 32, 414–423. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Wang, Z.; Xu, X.; Wan, Y.; Qu, K.; Fan, H.; Chen, Q.; Sun, X.; Liu, C. Nek7 is overexpressed in hepatocellular carcinoma and promotes hepatocellular carcinoma cell proliferation in vitro and in vivo. Oncotarget 2016, 7, 18620–18630. [Google Scholar] [CrossRef] [Green Version]
- Saloura, V.; Cho, H.-S.; Kiyotani, K.; Alachkar, H.; Zuo, Z.; Nakakido, M.; Tsunoda, T.; Seiwert, T.; Lingen, M.; Licht, J.; et al. WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol. Cancer Res. 2015, 13, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Eisa, N.H.; Jilani, Y.; Kainth, K.; Redd, P.; Lu, S.; Bougrine, O.; Abdul Sater, H.; Patwardhan, C.A.; Shull, A.; Shi, H.; et al. The co-chaperone UNC45A is essential for the expression of mitotic kinase NEK7 and tumorigenesis. J. Biol. Chem. 2019, 294, 5246–5260. [Google Scholar] [CrossRef] [PubMed]
- Vanaja, S.K.; Rathinam, V.A.K.; Atianand, M.K.; Kalantari, P.; Skehan, B.; Fitzgerald, K.A.; Leong, J.M. Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. USA 2014, 111, 7765–7770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanneganti, T.D.; Özören, N.; Body-Malapel, M.; Amer, A.; Park, J.H.; Franchi, L.; Whitfield, J.; Barchet, W.; Colonna, M.; Vandenabeele, P.; et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 2006, 440, 233–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, W.; Mitoma, H.; Hanabuchi, S.; Bao, M.; Weng, L.; Sugimoto, N.; Liu, Y.; Zhang, Z.; Zhong, J.; Sun, B.; et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc. Natl. Acad. Sci. USA 2014, 111, 16059–16064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, L.; Eigenbrod, T.; Núñez, G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009, 183, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Pétrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, H.; Gris, D.; Lei, Y.; Jha, S.; Zhang, L.; Huang, M.T.; Brickey, W.J.; Ting, J.P. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011, 12, 408–415. [Google Scholar] [CrossRef] [Green Version]
- Mathur, A.; Hayward, J.A.; Man, S.M. Molecular mechanisms of inflammasome signaling. J. Leukoc. Biol. 2018, 103, 233–257. [Google Scholar] [CrossRef]
- Pétrilli, V.; Papin, S.; Dostert, C.; Mayor, A.; Martinon, F.; Tschopp, J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007, 14, 1583–1589. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 103–109. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S.; et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016, 17, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, X.; Hu, Z.; Ye, X.; Zheng, X.; Ding, Y.; Xie, P.; Liu, Q. Protection of Mcc950 against high-glucose-induced human retinal endothelial cell dysfunction. Cell Death Dis. 2017, 8, e2941. [Google Scholar] [CrossRef]
- Ma, Z.-Z.; Sun, H.-S.; Lv, J.-C.; Guo, L.; Yang, Q.-R. Expression and clinical significance of the NEK7-NLRP3 inflammasome signaling pathway in patients with systemic lupus erythematosus. J. Inflamm. (Lond.) 2018, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, P.; Wang, Q.; Ji, N.; Xia, S.; Ding, Y.; Wang, Q. Metformin ameliorates experimental diabetic periodontitis independently of mammalian target of rapamycin (mTOR) inhibition by reducing NIMA-related kinase 7 (Nek7) expression. J. Periodontol. 2019, 90, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Mahjoub, M.R.; Trapp, M.L.; Quarmby, L.M. NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J. Am. Soc. Nephrol. 2005, 16, 3485–3489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grampa, V.; Delous, M.; Zaidan, M.; Odye, G.; Thomas, S.; Elkhartoufi, N.; Filhol, E.; Niel, O.; Silbermann, F.; Lebreton, C.; et al. Novel NEK8 Mutations Cause Severe Syndromic Renal Cystic Dysplasia through YAP Dysregulation. PLoS Genet. 2016, 12, e1005894. [Google Scholar] [CrossRef]
- Ding, X.F.; Chen, J.; Zhou, J.; Chen, G.; Wu, Y.L. Never-in-mitosis a-related kinase 8, a novel targof von-hippel-lindau tumor suppressor protein, promotes gastric cancer cell proliferation. Oncol. Lett. 2018, 16, 5900–5906. [Google Scholar]
- Ding, X.-F.; Zhou, J.; Hu, Q.-Y.; Liu, S.-C.; Chen, G. The tumor suppressor pVHL down-regulates never-in-mitosis A-related kinase 8 via hypoxia-inducible factors to maintain cilia in human renal cancer cells. J. Biol. Chem. 2015, 290, 1389–1394. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Molinari, E.; Raman, S.; Sayer, J.A. Many genes-one disease? Genetics of nephronophthisis (NPHP) and NPHP-associated disorders. Front. Pediatr. 2018, 5, 287. [Google Scholar] [CrossRef]
- Chaki, M.; Airik, R.; Ghosh, A.K.; Giles, R.H.; Chen, R.; Slaats, G.G.; Wang, H.; Hurd, T.W.; Zhou, W.; Cluckey, A.; et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 2012, 150, 533–548. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Otto, E.A.; Cluckey, A.; Airik, R.; Hurd, T.W.; Chaki, M.; Diaz, K.; Lach, F.P.; Bennett, G.R.; Gee, H.Y.; et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 2012, 44, 910–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, F. Exome resequencing identifies novel NPHP genes, implicating DNA damage response signaling in the pathogenesis of ciliopathies. Cilia 2012, 1, O2. [Google Scholar] [CrossRef] [Green Version]
- Abeyta, A.; Castella, M.; Jacquemont, C.; Taniguchi, T. NEK8 regulates DNA damage-induced RAD51 foci formation and replication fork protection. Cell Cycle 2017, 16, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Roig, J.; Mikhailov, A.; Belham, C.; Avruch, J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002, 16, 1640–1658. [Google Scholar] [CrossRef] [Green Version]
- Sdelci, S.; Schütz, M.; Pinyol, R.; Bertran, M.T.; Regué, L.; Caelles, C.; Vernos, I.; Roig, J. Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 2012, 22, 1516–1523. [Google Scholar] [CrossRef] [Green Version]
- O’Regan, L.; Barone, G.; Adib, R.; Woo, C.G.; Jeong, H.J.; Richardson, E.L.; Richards, M.W.; Muller, P.A.J.; Collis, S.J.; Fennell, D.A.; et al. EML4-ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7. J. Cell Sci. 2020. [Google Scholar] [CrossRef]
- Fathi, A.-R.; Roelcke, U. Meningioma. Curr. Neurol. Neurosci. Rep. 2013, 13, 337. [Google Scholar] [CrossRef]
- Dunn, J.; Ferluga, S.; Sharma, V.; Futschik, M.; Hilton, D.A.; Adams, C.L.; Lasonder, E.; Hanemann, C.O. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 2019, 40, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Krakow, D.; Geffen, D.; Author, C.P. Skeletal Dysplasias HHS Public Access Author manuscript. Clin. Perinatol. 2015, 42, 301–319. [Google Scholar] [CrossRef] [Green Version]
- Casey, J.P.; Brennan, K.; Scheidel, N.; McGettigan, P.; Lavin, P.T.; Carter, S.; Ennis, S.; Dorkins, H.; Ghali, N.; Blacque, O.E.; et al. Recessive NEK9 mutation causes a lethal skeletal dysplasia with evidence of cell cycle and ciliary defects. Hum. Mol. Genet. 2016, 25, 1824–1835. [Google Scholar] [CrossRef] [Green Version]
- Guldbakke, K.K.; Khachemoune, A.; Deng, A.; Sina, B. Naevus comedonicus: A spectrum of body involvement. Clin. Exp. Dermatol. 2007, 32, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Levinsohn, J.L.; Sugarman, J.L.; McNiff, J.M.; Antaya, R.J.; Choate, K.A. Somatic Mutations in NEK9 Cause Nevus Comedonicus. Am. J. Hum. Genet. 2016, 98, 1030–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, A.H.; Nelson, W.J.; Weis, W.I. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 1997, 90, 871–882. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Wu, D.; Hirao, A.; Lahti, J.M.; Liu, L.; Mazza, B.; Kidd, V.J.; Mak, T.W.; Ingram, A.J. ERK activation mediates cell cycle arrest and apoptosis after DNA damage independently of p53. J. Biol. Chem. 2002, 277, 12710–12717. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Black, C.P.; Cowan, K.H. Irradiation-induced G2/M checkpoint response requires ERK1/2 activation. Oncogene 2007, 26, 4689–4698. [Google Scholar] [CrossRef] [Green Version]
- Kaji, T.; Yamasaki, O.; Takata, M.; Otsuka, M.; Hamada, T.; Morizane, S.; Asagoe, K.; Yanai, H.; Hirai, Y.; Umemura, H.; et al. Comparative study on driver mutations in primary and metastatic melanomas at a single Japanese institute: A clue for intra- and inter-tumor heterogeneity. J. Dermatol. Sci. 2017, 85, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Ardila, D.E.; Ruigrok-Ritstier, K.; Helmijr, J.C.; Look, M.P.; van Laere, S.; Dirix, L.; Berns, E.M.J.J.; Jansen, M.P.H.M. LRG1 mRNA expression in breast cancer associates with PIK3CA genotype and with aromatase inhibitor therapy outcome. Mol. Oncol. 2016, 10, 1363–1373. [Google Scholar] [CrossRef] [Green Version]
- Milne, R.L.; Burwinkel, B.; Michailidou, K.; Arias-Perez, J.-I.; Zamora, M.P.; Menéndez-Rodríguez, P.; Hardisson, D.; Mendiola, M.; González-Neira, A.; Pita, G.; et al. Common non-synonymous SNPs associated with breast cancer susceptibility: Findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2014, 23, 6096–6111. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, K.; Fukazawa, H.; Murakami, Y.; Uehara, Y. Nucleolar Nek11 is a novel target of Nek2A in G1/S-arrested cells. J. Biol. Chem. 2004, 279, 32716–32727. [Google Scholar] [CrossRef] [Green Version]
- Sabir, S.R.; Sahota, N.K.; Jones, G.D.D.; Fry, A.M. Loss of Nek11 Prevents G2/M Arrest and Promotes Cell Death in HCT116 Colorectal Cancer Cells Exposed to Therapeutic DNA Damaging Agents. PLoS ONE 2015, 10, e0140975. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gao, Y.; Lu, Y.; Zhang, J.; Li, L.; Yin, F. Downregulation of NEK11 is associated with drug resistance in ovarian cancer. Int. J. Oncol. 2014, 45, 1266–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolf, M.G.; Curwen, J.O.; Veldman-Jones, M.; Eberlein, C.; Wang, J.; Harmer, A.; Hellawell, C.J.; Braddock, M. In vitro pharmacological profiling of R406 identifies molecular targets underlying the clinical effects of fostamatinib. Pharmacol. Res. Perspect. 2015, 3, e00175. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanfar, M.A.; Banat, F.; Alabed, S.; Alqtaishat, S. Discovery of potent NEK2 inhibitors as potential anticancer agents using structure-based exploration of NEK2 pharmacophoric space coupled with QSAR analyses. Mol. Divers. 2017, 21, 187–200. [Google Scholar] [CrossRef]
- Xi, J.-B.; Fang, Y.-F.; Frett, B.; Zhu, M.-L.; Zhu, T.; Kong, Y.-N.; Guan, F.-J.; Zhao, Y.; Zhang, X.-W.; Li, H.-Y.; et al. Structure-based design and synthesis of imidazo[1,2-a]pyridine derivatives as novel and potent Nek2 inhibitors with in vitro and in vivo antitumor activities. Eur. J. Med. Chem. 2017, 126, 1083–1106. [Google Scholar] [CrossRef]
- Ramachandran, B.; Kesavan, S.; Rajkumar, T. Molecular modeling and docking of small molecule inhibitors against NEK2. Bioinformation 2016, 12, 62–68. [Google Scholar] [CrossRef] [Green Version]
- De Donato, M.; Righino, B.; Filippetti, F.; Battaglia, A.; Petrillo, M.; Pirolli, D.; Scambia, G.; De Rosa, M.C.; Gallo, D. Identification and antitumor activity of a novel inhibitor of the NIMA-related kinase NEK6. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Musi, N.; Hirshman, M.F.; Nygren, J.; Svanfeldt, M.; Bavenholm, P.; Rooyackers, O.; Zhou, G.; Williamson, J.M.; Ljunqvist, O.; Efendic, S.; et al. Metformin increases AMP-activated protein-kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes 2002, 51, 2074–2081. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, E. Metformin-Induced Mitochondrial Complex I Inhibition: Facts, Uncertainties, and Consequences. Front. Endocrinol. (Lausanne) 2018, 9, 753. [Google Scholar] [CrossRef]
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014, 510, 542–546. [Google Scholar] [CrossRef] [Green Version]
- Meta, E.; Imhof, B.A.; Ropraz, P.; Fish, R.J.; Brullo, C.; Bruno, O.; Sidibé, A. The pyrazolyl-urea GeGe3 inhibits tumor angiogenesis and reveals dystrophia myotonica protein kinase (DMPK)1 as a novel angiogenesis target. Oncotarget 2017, 8, 108195–108212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, J.; Stenger, M. Dabrafenib in advanced melanoma with BRAF V600E mutation. J. Community Support. Oncol. 2014, 12, 48–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibney, G.T.; Zager, J.S. Clinical development of dabrafenib in BRAF mutant melanoma and other malignancies. Expert Opin. Drug Metab. Toxicol. 2013, 9, 893–899. [Google Scholar] [CrossRef] [PubMed]



| Drug Name | Chemical Formula | NEK Targets | Action/Effects | Stage of Development | Selectivity | References |
|---|---|---|---|---|---|---|
| Fostamatinib | C23H26FN6O9P | NEK1 NEK3 NEK4 NEK5 NEK9 NEK11 | Inhibits signal transduction by Fcγ receptors involved in the antibody-mediated destruction of platelets. NEKs 1,2,3,4 5,9,11 are included in the list of targets | Approved for the treatment of chronic immune thrombocytopenia (ITP). Under investigation for other diseases | The active metabolite of fostamatinib, named R406, presents a lack of selectivity among several kinases | [203] |
| 5-[(Z)-(5-Chloro-2-oxo-1,2-dihydro-3H-indol-3-ylidene) methyl]-N,2,4-trimethyl-1H-pyrrole-3-carboxamide | C17H16ClN3O2 | NEK2 | Interacts with NEK2 | Experimental | No off-targets have been described | [204] |
| NCI code 51,525 and 58991 | - | NEK2 | Inhibits NEK2 | Experimental | No off-targets have been described | [205] |
| MBM-17 and MBM-55 | C28H29N6 and C28H28N6O2F respectively | NEK2 | Inhibits NEK2 and has antiproliferative/antitumor properties | Experimental | It has low nanomolar activity and a great selectivity for NEK2 | [206] |
| di-demethylchlorpromazine and 2-[5-fluoro-1H-indol-3-yl] propan-1-amine | C15-H15-Cl-N2-S.Cl-H and C11H14ClFN2 respectively | NEK2 | Inhibits NEK2 | Pre-experimental (in silico) | No off-targets have been described | [207] |
| (5Z)-2-hydroxy-4-methyl-6-oxo-5-[(5-phenylfuran-2-yl)methylidene]-5,6-dihydropyridine-3-carbonitrile) or compound 8 | C22H24F3N8O2 | NEK6 NEK1 | Inhibits NEK6. Displays anti-proliferative effects on several cancer cell lines with low IC50 values. Induces cell cycle arrest in the G2/M phases and has a synergistic effect with cisplatin and paclitaxel in ovarian cancer. It can inhibit NEK1. | Experimental | Inhibits NEK6 and NEK1. No off-targets have been described | [208] |
| Metformin | C4H11N5 | NEK7 | Metformin inhibits NEK7 expression in an experimental diabetic periodontitis model | Approved for type II diabetes | Activates AMPK, inhibits electron transfer flavoprotein-ubiquinone oxidoreductase (ETFDH), and glycerol-3-phosphate dehydrogenase [NAD(+)] (GPD1) | [175,209,210,211] |
| Ethyl 1-(2-hydroxypentyl) 5-(3-(3-(trifluoromethyl) phenyl)ureido)-1H-pyrazole-4-carboxylate or GeGe3 | - | NEK10 | Screening for GeGe3-targeted kinases revealed NEK10 as candidate targets, such as Aurora B. Inhibits physiological and tumor angiogenesis. | Experimental | Strongly inhibits aurora B, aurora C, NEK 10, polo like kinases 2 and 3 (PLK2/PLK3), dystrophia myotonica protein kinase 1 (DMPK), and calcium/calmodulin-dependent protein kinase type 1 (CaMK1) | [212] |
| Dabrafenib | C23H20F3N5O2S2 | NEK11 | Reduces the proliferation and regression of tumors in xenograft models. | Approved for the treatment of metastatic melanoma with BRAF V600E mutations | Inhibits serine/threonine-protein kinase B-raf (BRAF1), proto-oncogene c-RAF (RAF), serine/threonine-protein kinase SIK1 (SIK-1), NEK11, and LIM domain kinase 1 (LIMK-1) | [213,214] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peres de Oliveira, A.; Kazuo Issayama, L.; Betim Pavan, I.C.; Riback Silva, F.; Diniz Melo-Hanchuk, T.; Moreira Simabuco, F.; Kobarg, J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules 2020, 25, 1778. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25081778
Peres de Oliveira A, Kazuo Issayama L, Betim Pavan IC, Riback Silva F, Diniz Melo-Hanchuk T, Moreira Simabuco F, Kobarg J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules. 2020; 25(8):1778. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25081778
Chicago/Turabian StylePeres de Oliveira, Andressa, Luidy Kazuo Issayama, Isadora Carolina Betim Pavan, Fernando Riback Silva, Talita Diniz Melo-Hanchuk, Fernando Moreira Simabuco, and Jörg Kobarg. 2020. "Checking NEKs: Overcoming a Bottleneck in Human Diseases" Molecules 25, no. 8: 1778. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25081778