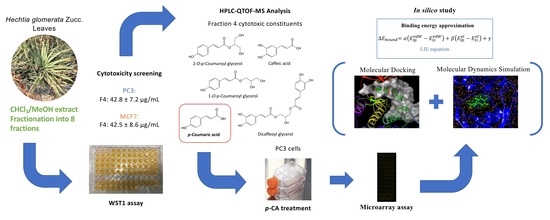
Cytotoxic Fractions from Hechtia glomerata Extracts and p-Coumaric Acid as MAPK Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Plant Material Extraction
2.2. Cytotoxic Activity of Extracts
2.3. Cytotoxic Screening of Pooled Fractions
2.4. HPLC-QTOF-MS Analysis of Cytotoxic Fraction 4
2.5. Cytotoxicity of p-CA
2.6. Microarray Assay
2.7. Molecular Docking and Molecular Dynamics Simulations
3. Materials and Methods
3.1. Plant Material
3.2. Extract Preparation
3.3. Fractionation of CHCl3/MeOH Extract by Column Chromatography
3.4. Cell Lines and Cytotoxicity Assay
3.5. HPLC-QTOF-MS Analysis
3.6. Cell Treatment and Isolation of Total RNA
3.7. Microarray Chip Printing
3.8. Labeled cDNA Synthesis and Microarray Assay
3.9. Microarray Data Analysis
3.10. Molecular Docking
3.11. Molecular Dynamics Simulations
3.12. Binding Energy Approximation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Shakya, A.K. Medicinal plants: Future source of new drugs. Int. J. Herb. Med. 2016, 4, 59–64. [Google Scholar]
- Hornung-Leoni, C.T. Avances sobre usos etnobotánicos de las Bromeliaceae en Latinoamérica. Bol. Latinoam. Caribe Plantas Med. Aromat. 2011, 10, 297–314. [Google Scholar]
- Sandoval-Bucio, E.N.; Flores-Cruz, M.; Martinez-Bernal, A. Bromelias útiles en México (Useful bromeliads in Mexico). Cactáceas Suculentas Mex. 2004, 49, 100–115. [Google Scholar]
- Givnish, T.J.; Barfuss, M.H.J.; Van Ee, B.; Riina, R.; Schulte, K.; Horres, R.; Gonsiska, P.A.; Jabaily, R.S.; Crayn, D.M.; Smith, J.A.C.; et al. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: Insights from an eight-locus plastid phylogeny. Am. J. Bot. 2011, 98, 872–895. [Google Scholar] [CrossRef] [Green Version]
- Lowe, H.; Watson, C.T.; Badal, S.; Toyang, N.J.; Bryant, J. Unearthing the Medicinal Properties of Tillandsia recurvata (Ball Moss): A Mini Review. Eur. J. Med. Plants 2014, 4, 1138–1149. [Google Scholar] [CrossRef] [Green Version]
- Lowe, H.I.; Toyang, N.J.; Watson, C.; Badal, S.; Bahado-Singh, P.; Bryant, J. In Vitro Anticancer Activity of the Crude Extract and two Dicinnamate Isolates from the Jamaican Ball Moss (Tillandsia Recurvata L.). Am. Int. J. Contemp. Res. 2013, 3, 93–96. [Google Scholar]
- Lowe, H.I.; Toyang, N.J.; Watson, C.T.; Ayeah, K.N.; Bryant, J. Antileukemic activity of Tillandsia recurvata and some of its cycloartanes. Anticancer. Res. 2014, 34, 3505–3509. [Google Scholar]
- Cos, P.; Vlietinck, A.J.; Berghe, D.V.; Maes, L. Anti-infective potential of natural products: How to develop a stronger in vitro ‘proof-of-concept’. J. Ethnopharmacol. 2006, 106, 290–302. [Google Scholar] [CrossRef]
- Stefani, T.; Garza-González, E.; Rivas-Galindo, V.M.; Rios, M.Y.; Alvarez, L.; Camacho-Corona, M.d.R. Hechtia glomerata Zucc: Phytochemistry and Activity of Its Extracts and Major Constituents Against Resistant Bacteria. Molecules 2019, 24, 3434. [Google Scholar] [CrossRef] [Green Version]
- Stefani, T.; Claudio, P.D.C.M.-S.; Rios, M.Y.; Aguilar-Guadarrama, A.B.; González-Maya, L.; Sánchez-Carranza, J.N.; González-Ferrara, M.; Camacho-Corona, M.D.R. UPLC–QTOF–MS analysis of cytotoxic and antibacterial extracts of Hechtia glomerata Zucc. Nat. Prod. Res. 2020, 1–5. [Google Scholar] [CrossRef]
- Pei, K.; Ou, J.; Huang, J.; Ou, S. p-Coumaric acid and its conjugates: Dietary sources, pharmacokinetic properties and biological activities. J. Sci. Food Agric. 2016, 96, 2952–2962. [Google Scholar] [CrossRef]
- Bouzaiene, N.N.; Jaziri, S.K.; Kovacic, H.; Chekir-Ghedira, L.; Ghedira, K.; Luis, J. The effects of caffeic, coumaric and ferulic acids on proliferation, superoxide production, adhesion and migration of human tumor cells in vitro. Eur. J. Pharmacol. 2015, 766, 99–105. [Google Scholar] [CrossRef]
- Shailasree, S.; Venkataramana, M.; Niranjana, S.R.; Prakash, H.S. Cytotoxic Effect of p-Coumaric Acid on Neuroblastoma, N2a Cell via Generation of Reactive Oxygen Species Leading to Dysfunction of Mitochondria Inducing Apoptosis and Autophagy. Mol. Neurobiol. 2015, 51, 119–130. [Google Scholar] [CrossRef]
- Turpaev, K.T. Reactive Oxygen Species and Regulation of Gene Expression. Biochemistry (Moscow) 2002, 67, 281–292. [Google Scholar] [CrossRef]
- Grever, M.R.; A Schepartz, S.; A Chabner, B. The National Cancer Institute: Cancer drug discovery and development program. Semin. Oncol. 1992, 19, 622–638. [Google Scholar]
- Nguyen, N.H.; Ta, Q.T.H.; Pham, Q.T.; Luong, T.N.H.; Phung, V.T.; Duong, T.-H.; Vo, V.G. Anticancer Activity of Novel Plant Extracts and Compounds from Adenosma bracteosum (Bonati) in Human Lung and Liver Cancer Cells. Molecules 2020, 25, 2912. [Google Scholar] [CrossRef]
- Kang, J.; Price, W.E.; Ashton, J.; Tapsell, L.C.; Johnson, S. Identification and characterization of phenolic compounds in hydromethanolic extracts of sorghum wholegrains by LC-ESI-MSn. Food Chem. 2016, 211, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Xiao, S.-Y.; Li, Z.-G.; Wang, W.; Du, L.-J. Characterization of active phenolic components in the ethanolic extract of Ananas comosus L. leaves using high-performance liquid chromatography with diode array detection and tandem mass spectrometry. J. Chromatogr. A 2007, 1165, 39–44. [Google Scholar] [CrossRef]
- Steingass, C.B.; Glock, M.P.; Schweiggert, R.M.; Carle, R. Studies into the phenolic patterns of different tissues of pineapple (Ananas comosus [L.] Merr.) infructescence by HPLC-DAD-ESI-MS n and GC-MS analysis. Anal. Bioanal. Chem. 2015, 407, 6463–6479. [Google Scholar] [CrossRef] [PubMed]
- Estrella-Parra, E.; Flores-Cruz, M.; Blancas-Flores, G. The Tillandsia genus: History, uses, chemistry, and biological activity. Bol. Latinoam. Caribe Plantas Med. Aromát. 2019, 18, 239–264. [Google Scholar]
- Manetti, L.M.; Deiaporte, R.H.; Laverde, A. Metabólitos secundários da família bromeliaceae. Quim. Nova 2009, 32, 1885–1897. [Google Scholar]
- Lowe, H.I.C.; Watson, C.T.; Badal, S.; Toyang, N.J.; Bryant, J. Kinase inhibition by the Jamaican ball moss, Tillandsia recurvata L. Anticancer. Res. 2012, 32, 4419–4422. [Google Scholar]
- Reddivari, L.; Vanamala, J.; Safe, S.H.; Miller, C., Jr. The Bioactive Compounds α-Chaconine and Gallic Acid in Potato Extracts Decrease Survival and Induce Apoptosis in LNCaP and PC3 Prostate Cancer Cells. Nutr. Cancer 2010, 62, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Hanham, A.F.; Dunn, B.P.; Stich, H.F. Clastogenic activity of caffeic acid and its relationship to hydrogen peroxide generated during autooxidation. Mutat. Res. Toxicol. 1983, 116, 333–339. [Google Scholar] [CrossRef]
- Lin, H.-P.; Jiang, S.S.; Chuu, C.-P. Caffeic Acid Phenethyl Ester Causes p21Cip1 Induction, Akt Signaling Reduction, and Growth Inhibition in PC-3 Human Prostate Cancer Cells. PLoS ONE 2012, 7, e31286. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, K.; Singh, S.; Burke, T.R.; Grunberger, D.; Aggarwal, B.B. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proc. Natl. Acad. Sci. USA 1996, 93, 9090–9095. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Shang, M.-Y.; Liu, G.-X.; Xu, F.; Wang, X.; Shou, C.-C.; Cai, S.-Q. Chemical Constituents from the Rhizomes of Smilax glabra and Their Antimicrobial Activity. Molecules 2013, 18, 5265–5287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaporte, R.H.; Guzen, K.P.; LaVerde, A.; Dos Santos, A.R.; Sarragiotto, M.H. Phenylpropanoid glycerols from Tillandsia streptocarpa Baker (Bromeliaceae). Biochem. Syst. Ecol. 2006, 34, 599–602. [Google Scholar] [CrossRef]
- Russo, G.L.; Russo, M.; Spagnuolo, C.; Tedesco, I.; Bilotto, S.; Iannitti, R.; Palumbo, R. Quercetin: A Pleiotropic Kinase Inhibitor Against Cancer. Cancer Treat. Res. 2013, 159, 185–205. [Google Scholar] [CrossRef]
- Park, S.E.; Sapkota, K.; Kim, S.J.; Kim, H. Kaempferol acts through mitogen-activated protein kinases and protein kinase B/AKT to elicit protection in a model of neuroinflammation in BV2 microglial cells. Br. J. Pharmacol. 2011, 164, 1008–1025. [Google Scholar] [CrossRef] [Green Version]
- Hashemzaei, M.; Far, A.D.; Yari, A.; Heravi, R.E.; Tabrizian, K.; Taghdisi, S.M.; Sadegh, S.E.; Tsarouhas, K.; Kouretas, D.; Tzanakakis, G.; et al. Anticancer and apoptosis-inducing effects of quercetin in vitro and in vivo. Oncol. Rep. 2017, 38, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.W.; Kang, N.J.; Heo, Y.-S.; Rogozin, E.A.; Pugliese, A.; Hwang, M.K.; Bowden, G.T.; Bode, A.M.; Lee, H.J.; Dong, Z. Raf and MEK Protein Kinases Are Direct Molecular Targets for the Chemopreventive Effect of Quercetin, a Major Flavonol in Red Wine. Cancer Res. 2008, 68, 946–955. [Google Scholar] [CrossRef] [Green Version]
- Anwar, S.; Shamsi, A.; Shahbaaz, M.; Queen, A.; Khan, P.; Hasan, G.M.; Islam, A.; Alajmi, M.F.; Hussain, A.; Ahmad, F.; et al. Rosmarinic Acid Exhibits Anticancer Effects via MARK4 Inhibition. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Brack, E. Exploiting Oxidative Stress as a Therapeutic Option in Rhabdomyosarcoma. Ph.D. Thesis, University of Zurich, Zurich, Switzerland, 2018.
- Wagner, T.M.; Mullally, J.E.; Fitzpatrick, F.A. Reactive lipid species from cyclooxygenase-2 inactivate tumor suppressor LKB1/STK11 cyclopentenone prostaglandins and 4-hydroxy-2-nonenal covalently modify and inhibit the amp-kinase kinase that modulates cellular energy homeostasis and protein translatio. J. Biol. Chem. 2006, 281, 2598–2604. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.; Zhong, Z.; Carney, R.P.; Men, Y.; Li, J.; Pan, T.; Wang, Y. Deciphering the metabolic role of AMPK in cancer multi-drug resistance. Semin. Cancer Biol. 2019, 56, 56–71. [Google Scholar] [CrossRef]
- Chang, W.H.; Lai, A.G. An integrative pan-cancer investigation reveals common genetic and transcriptional alterations of AMPK pathway genes as important predictors of clinical outcomes across major cancer types. BMC Cancer 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Lee, S.; Rauch, J.; Kolch, W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci. 2020, 21, 1102. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Spigolon, G.; Bonny, C.; Culman, J.; Vercelli, A.; Herdegen, T. The JNK inhibitor D-JNKI-1 blocks apoptotic JNK signaling in brain mitochondria. Mol. Cell. Neurosci. 2012, 49, 300–310. [Google Scholar] [CrossRef]
- Stebbins, J.L.; De, S.K.; Machleidt, T.; Becattini, B.; Vazquez, J.; Kuntzen, C.; Chen, L.-H.; Cellitti, J.F.; Riel-Mehan, M.; Emdadi, A.; et al. Identification of a new JNK inhibitor targeting the JNK-JIP interaction site. Proc. Natl. Acad. Sci. USA 2008, 105, 16809–16813. [Google Scholar] [CrossRef] [Green Version]
- Cenda, A. Mitogen-activated protein kinases (MAPK) in cancer. In Encyclopedia of Cancer, 3rd ed.; Boffetta, P., Hainaut, P., Eds.; Elsevier: New York, NY, USA, 2019; pp. 472–480. [Google Scholar]
- Li, Z.; Sun, C.; Tao, S.; Osunkoya, A.O.; Arnold, R.S.; Petros, J.A.; Zu, X.; Moreno, C.S. The JNK inhibitor AS602801 Synergizes with Enzalutamide to Kill Prostate Cancer Cells In Vitro and In Vivo and Inhibit Androgen Receptor Expression. Transl. Oncol. 2020, 13, 100751. [Google Scholar] [CrossRef]
- Baek, S.; Kang, N.J.; Popowicz, G.M.; Arciniega, M.; Jung, S.K.; Byun, S.; Song, N.R.; Heo, Y.-S.; Kim, B.Y.; Lee, H.J.; et al. Structural and functional analysis of the natural JNK1 inhibitor quercetagetin. J. Mol. Biol. 2013, 425, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.-J.; Li, X.; Wang, W.; Zi, X.; Zhang, R. Targeting the NFAT1-MDM2-MDMX Network Inhibits the Proliferation and Invasion of Prostate Cancer Cells, Independent of p53 and Androgen. Front. Pharmacol. 2017, 8, 917. [Google Scholar] [CrossRef] [Green Version]
- Lenos, K.; Jochemsen, A.G. Functions of MDMX in the Modulation of the p53-Response. J. Biomed. Biotechnol. 2011, 2011, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Xiang, P.; Sun, Y.; Fang, Z.; Yan, K.; Fan, Y. Eukaryotic translation initiation factor 3 subunit b is a novel oncogenic factor in prostate cancer. Mamm. Genome 2020, 31, 197–204. [Google Scholar] [CrossRef]
- Vaysse, C.; Philippe, C.; Martineau, Y.; Quelen, C.; Hieblot, C.; Renaud, C.; Nicaise, Y.; Desquesnes, A.; Pannese, M.; Filleron, T.; et al. Key contribution of eIF4H-mediated translational control in tumor promotion. Oncotarget 2015, 6, 39924–39940. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.-F.; Li, W.; Shi, F.; Zhou, Z.-W.; Li, B.; Zhang, K.; Zhang, X.; Ouyang, C.; Zhu, X. A bioinformatic and mechanistic study elicits the antifibrotic effect of ursolic acid through the attenuation of oxidative stress with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in human hepatic stellate cells and rat liver. Drug Des. Dev. Ther. 2015, 9, 3989–4104. [Google Scholar] [CrossRef] [Green Version]
- Qian, C.; Qi, Y.; Zhong, S.; Zeng, J.; Chen, X.; Yao, J. Mitogen-activated protein kinase inhibition enhances the antitumor effects of sporamin in human pancreatic cancer cells. Oncol. Lett. 2018, 16, 1237–1242. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Xin, Z.; Clampit, J.E.; Wang, S.; Gum, R.J.; Haasch, D.L.; Trevillyan, J.M.; Abad-Zapatero, C.; Fry, E.H.; Sham, H.L.; et al. Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors. Bioorganic Med. Chem. Lett. 2006, 16, 2590–2594. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, A.K.; Kushwaha, P.P.; Prajapati, K.S.; Shuaib, M.; Senapati, S.; Kumar, S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Raut, G.K.; Manchineela, S.; Chakrabarti, M.; Bhukya, C.K.; Naini, R.; Venkateshwari, A.; Reddy, V.; Mendonza, J.J.; Suresh, Y.; Nallari, P.; et al. Imine stilbene analog ameliorate isoproterenol-induced cardiac hypertrophy and hydrogen peroxide-induced apoptosis. Free Radic. Biol. Med. 2020, 153, 80–88. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Kumar, A.; Maurya, S.; Singh, A.K.; Sharma, A.K.; Kumar, S. Bulbine frutescens Phytochemicals as a Promising Anti-cancer Drug Discovery Source: A Computational Study. In Phytochemistry: An in-silico and in-vitro Update; Kumar, S., Egbuna, C., Eds.; Springer International Publishing: Singapore, 2019; pp. 491–510. [Google Scholar]
- Huang, S.-Y.; Bolser, D.; Liu, H.-Y.; Hwang, T.-C.; Zou, X. Molecular modeling of the heterodimer of human CFTR’s nucleotide-binding domains using a protein–protein docking approach. J. Mol. Graph. Model. 2009, 27, 822–828. [Google Scholar] [CrossRef] [Green Version]
- Basu, A.; Saito, K.; Meyer, K.; Ray, R.B.; Friedman, S.L.; Chang, Y.-H.; Ray, R. Stellate cell apoptosis by a soluble mediator from immortalized human hepatocytes. Apoptosis 2006, 11, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Espinoza, E.U.; López-Cortina, S.T.; Ramírez-Cabrera, M.A.; Balderas-Rentería, I. Synthesis and photodynamic activity of unsymmetrical A3B tetraarylporphyrins functionalized with l-glutamate and their Zn(II) and Cu(II) metal complex derivatives. Biomed. Pharmacother. 2016, 82, 327–336. [Google Scholar] [CrossRef]
- Clemente-Soto, A.F.; Balderas-Rentería, I.; Rivera, G.; Segura-Cabrera, A.; Garza-González, E.; Camacho-Corona, M.D.R. Potential Mechanism of Action of meso-Dihydroguaiaretic Acid on Mycobacterium tuberculosis H37Rv. Molecules 2014, 19, 20170–20182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Shin, W.H.; Lee, G.R.; Heo, L.; Lee, H.; Seok, C. Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio Des. 2014, 2, 1–11. [Google Scholar]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Shapovalov, M.V.; Dunbrack, R.L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511. [Google Scholar] [CrossRef] [Green Version]
- Kassir, Y.; Kupiec, M.; Shalom, A.; Simchen, G. Cloning and mapping of CDC40, a Saccharomyces cerevisiae gene with a role in DNA repair. Curr. Genet. 1985, 9, 253–257. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Andoh, Y.; Yoshii, N.; Yamada, A.; Okazaki, S. Evaluation of atomic pressure in the multiple time-step integration algorithm. J. Comput. Chem. 2017, 38, 704–713. [Google Scholar] [CrossRef]
- Aqvist, J.; Marelius, J. The linear interaction energy method for predicting ligand binding free energies. Comb. Chem. High Throughput Screen. 2001, 4, 613–626. [Google Scholar] [CrossRef]



| Extract | IC50 a ± SD b (µg/mL) | |
|---|---|---|
| PC3 c | MCF7 d | |
| Hexane | 76 ± 5 | 27 ± 2.5 |
| C/M e | 25 ± 4 | 32 ± 2 |
| Aqueous | 81 ± 3 | 71 ± 3 |
| Positive control | IC50 ± SD (µg/mL) | |
| Paclitaxel | 13.2 ± 3 × 10−3 | 33.7 ± 3.9 × 10−3 |
| Pooled Fraction (g) | IC50 (µg/mL) | |
|---|---|---|
| MCF7 | PC3 | |
| 1 (1.07) | >100 | >100 |
| 2 (11.09) | >100 | 97.4 ± 14.8 |
| 3 (18.30) | 99.5 ± 7.3 | 84.1 ± 8.4 |
| 4 (9.60) | 42.5 ± 8.6 | 42.8 ± 7.2 |
| 5 (6.78) | >100 | >100 |
| 6 (4.98) | >100 | >100 |
| 7 (7.38) | >100 | >100 |
| 8 (12.70) | >100 | >100 |
| Positive control | IC50 (µg/mL) | |
| Paclitaxel | 33.7 ± 3.9 × 10−3 | 13.2 ± 3 × 10−3 |
| Retention Time (min) | Compound | Calculated Formula | [M − H]− | [M + H]+ | Fragments | Area (%) | Ref. |
|---|---|---|---|---|---|---|---|
| 1.001 | Protocatechuic acid | C7H6O4 | 153.0189 | - | [M − H]−: 109 | 0.89 | [18] |
| 1.732 | Salicylic acid | C7H6O3 | 137.0239 | - | [M − H]−: 93 | 0.86 | a, b |
| 3.129 | Caffeic acid | C9H8O4 | 179.0342 | - | [M − H]−: 135 | 0.24 | a |
| 4.416 | 2-O-p-Coumaroyl glycerol | C12H14O5 | 237.0768 | 239.0900 | [M − H]−: 119, 145, 163 [M + H]+: 147, 119 | 7.22 | [18] |
| 4.737 | p-Coumaric acid | C9H8O3 | 163.0400 | - | [M − H]−: 119 | 6.05 | [19] |
| 4.99 | 1-O-p-Coumaroyl glycerol | C12H14O5 | 237.0767 | - | [M − H]−: 119, 145, 163 | 12.18 | c |
| 5.384 | Unknown | NA | - | 275.1300 | [M + H]+: 79, 197, 179, 135 | 3.84 | m |
| 6.561 | Dicaffeoyl glycerol isomer | C21H20O9 | - | 417.1200 | [M + H]+: 163, 145, 237 | 3.50 | c |
| 6.741 | Dicaffeoyl glycerol | C21H20O9 | 415.1044 | 417.1200 | [M − H]−: 253, 161, 179, 135, 237 [M + H]+: 163, 145, 164, 237 | 14.53 | [20] |
| 7.063 | Caffeoyl coumaroyl glycerol | C21H20O8 | 399.1102 | 401.1200 | [M − H]−: 163, 253, 119, 145, 135, 235, 179 [M + H]+: 147, 163, 145, 119 | 3.66 | m |
| 7.514 | Naringenin derivative | C24H20O8 | - | 437.1200 | [M + H]+: 437, 177, 251, 147, 243, 221 | 2.22 | d |
| 7.569 | Unknown | C17H14O7 | - | 331.0800 | [M + H]+: 331, 315, 298, 273, 281 | 3.83 | m |
| 7.837 | Chlorophyll derivative | NA | - | 843.2300 | [M + H]+: 843, 555, 844, 597, 556 | 10.32 | d |
| 8.013 | Unknown | C19H18O9 | - | 391.1000 | [M + H]+: 391, 361, 389, 375, 358 | 2.70 | m |
| 8.014 | Coumarin isomer | C9H6O2 | - | 147.0400 | [M + H]+: 91, 119, 147, 65 | 1.84 | m |
| 8.168 | Coumarin derivative | C20H22O6 | - | 359.1500 | [M + H]+: 147, 119 | 5.32 | m |
| 8.215 | 3-(4-Hydroxyphenyl)propyl coumarate | C18H18O4 | 297.1151 | - | [M − H]−: 119, 145, 163 [M + H]+: 107, 147, 135 | 4.33 | m |
| 8.297 | 3-(4-Hydroxyphenyl-3-methoxy) propyl coumarate | C19H20O5 | 327.1255 | - | [M − H]−: 163, 119, 145, 177, 133, 312, 299 | 3.02 | m |
| 8.37 | Kaempferol or cyanidin derivative | C18H16O7 | - | 345.1000 | [M + H]+: 345, 312, 329, 330, 287 | 3.37 | d |
| 8.784 | Palmitoleic acid | C16H30O2 | - | 255.2300 | [M + H]+: 69, 67, 83, 55, 81, 97, 95, 71, 43, 135, 93, 121, 107, 97, 149, 67, 109 | 1.71 | m |
| 9.156 | Demethylnobiletin | C20H20O8 | - | 389.1200 | [M + H]+: 389, 356, 331, 373, 359, 374 | 5.74 | e |
| 9.453 | Syringaresinol | C21H22O9 | - | 419.1300 | [M + H]+: 419, 389, 371, 420, 390 | 2.63 | m |
| Compound | Biological Effects | IC50/MIC a | Target/Cells | Source | Ref. |
|---|---|---|---|---|---|
| 1-O-p-Coumaroyl glycerol | - | - | - | Ananas cumosus | [22,28] |
| 2-O-p-Coumaroyl glycerol | Antimicrobial | MRSA b | |||
| Caffeic acid | Cytotoxicity | 177.62 µM | PC3 | Purple potato extract | [24] |
| Caffeoyl coumaroyl glycerol | Cytotoxic effects | - | - | Tillandsia streptocarpa | [29] |
| Dicaffeoyl glycerol | - | - | |||
| p-Coumaric acid | ROS, anti-angiogenesis, AKT and ERK signaling pathways | >500 µM | MCF7, murine tumors, N2a, ECV304 | - | [13] |
| Functional Category | Upregulated Genes (%) | Downregulated Genes (%) |
|---|---|---|
| Acetylation | - | 23.73 |
| Activator | 9.13 | - |
| Apoptosis | 4.78 | - |
| ATP-binding | 10.87 | - |
| Calcium | 7.39 | - |
| Cell membrane | - | 22.60 |
| Coiled coil | 19.13 | - |
| Developmental protein | 8.70 | - |
| Differentiation | 6.52 | - |
| Disulfide bond | - | 23.16 |
| DNA damage | 3.48 | - |
| DNA-binding | 18.70 | - |
| EGF-like domain | 3.91 | - |
| G-protein coupled receptor | - | 10.17 |
| Kinase | - | 7.91 |
| Lipoprotein | 6.52 | - |
| Magnesium | - | 6.21 |
| Membrane | - | 45.76 |
| Metal-binding | 29.57 | 23.16 |
| Nucleotide-binding | 11.74 | - |
| Nucleus | 36.09 | - |
| Phosphoprotein | 45.65 | 55.37 |
| Proto-oncogene | - | 3.95 |
| Receptor | - | 15.25 |
| Repressor | 5.22 | - |
| Sugar transport | - | 1.69 |
| Transcription regulation | 22.17 | - |
| Transducer | - | 10.17 |
| Transferase | - | 14.12 |
| Transmembrane | - | 36.16 |
| Transport | - | 14.12 |
| Ubl conjugation | - | 14.69 |
| Zinc | 21.74 | - |
| Zinc-finger | 18.70 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefani, T.; Romo-Mancillas, A.; Carrizales-Castillo, J.J.J.; Arredondo-Espinoza, E.; Ramírez-Estrada, K.; Alcantar-Rosales, V.M.; González-Maya, L.; Sánchez-Carranza, J.N.; Balderas-Renterías, I.; Camacho-Corona, M.d.R. Cytotoxic Fractions from Hechtia glomerata Extracts and p-Coumaric Acid as MAPK Inhibitors. Molecules 2021, 26, 1096. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26041096
Stefani T, Romo-Mancillas A, Carrizales-Castillo JJJ, Arredondo-Espinoza E, Ramírez-Estrada K, Alcantar-Rosales VM, González-Maya L, Sánchez-Carranza JN, Balderas-Renterías I, Camacho-Corona MdR. Cytotoxic Fractions from Hechtia glomerata Extracts and p-Coumaric Acid as MAPK Inhibitors. Molecules. 2021; 26(4):1096. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26041096
Chicago/Turabian StyleStefani, Tommaso, Antonio Romo-Mancillas, Juan J. J. Carrizales-Castillo, Eder Arredondo-Espinoza, Karla Ramírez-Estrada, Victor M. Alcantar-Rosales, Leticia González-Maya, Jessica Nayelli Sánchez-Carranza, Isaías Balderas-Renterías, and María del Rayo Camacho-Corona. 2021. "Cytotoxic Fractions from Hechtia glomerata Extracts and p-Coumaric Acid as MAPK Inhibitors" Molecules 26, no. 4: 1096. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26041096