The Improved Milk Quality and Enhanced Anti-Inflammatory Effect in Acetylserotonin-O-methyltransferase (ASMT) Overexpressed Goats: An Association with the Elevated Endogenous Melatonin Production
Abstract
:1. Introduction
2. Results
2.1. Production and Identification of ASMT Overexpressed Dairy Goats
2.2. Determination of Melatonin Levels in Blood and Milk and Analysis of the Milk Quality
2.3. Microbiota Distribution Analysis
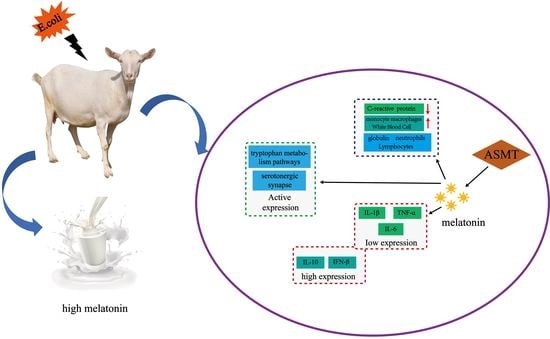
2.4. Changes in Body Physiological Indexes after E. coli Injection
2.5. Changes of Inflammatory Biomarkers
2.6. Serum Metabolism Analysis after E. coli Injection
2.7. ASMT Overexpression Associated Metabolic Consequences
3. Discussion
4. Materials and Methods
4.1. Construction of ASMT Overexpressed Vector
4.2. Prokaryotic Embryo Microinjection Procedure
4.3. Identification of Progeny
4.4. Melatonin Assay
4.5. Milk Quality Analysis
4.6. 16S rDNA Sequencing and Analysis
4.7. Colony-Forming Unit (CFU) Counts
4.8. Animal Study Design
4.9. Analysis of Physiological and Biochemical Indexes
4.10. Radioimmunoassay of Inflammatory Cytokines
4.11. Metabolome Analysis
4.12. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bhattacharya, S.; Patel, K.K.; Dehari, D.; Agrawal, A.K.; Singh, S. Melatonin and its ubiquitous anticancer effects. Mol. Cell. Biochem. 2019, 462, 133–155. [Google Scholar] [CrossRef] [PubMed]
- Borjigin, J.; Li, X.; Snyder, S.H. The pineal gland and melatonin: Molecular and pharmacologic regulation. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.X.; Reiter, R.J. Extrapineal melatonin: Sources, regulation, and potential functions. Cell. Mol. Life Sci. 2014, 71, 2997–3025. [Google Scholar] [CrossRef]
- Carrascal, L.; Nunez-Abades, P.; Ayala, A.; Cano, M. Role of Melatonin in the Inflammatory Process and Its Therapeutic Potential. Curr. Pharm. Des. 2018, 24, 1563–1588. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Perez, J.L.; Lopez-Patino, M.A.; Alvarez-Otero, R.; Gesto, M.; Soengas, J.L.; Miguez, J.M. Characterization of melatonin synthesis in the gastrointestinal tract of rainbow trout (Oncorhynchus mykiss): Distribution, relation with serotonin, daily rhythms and photoperiod regulation. J. Comp. Physiol. B 2016, 186, 471–484. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Nabavi, S.F.; Sureda, A.; Xiao, J.; Dehpour, A.R.; Shirooie, S.; Silva, A.S.; Baldi, A.; Khan, H.; Daglia, M. Anti-inflammatory effects of Melatonin: A mechanistic review. Crit. Rev. Food Sci. Nutr. 2019, 59, S4–S16. [Google Scholar] [CrossRef]
- Shukla, M.; Govitrapong, P.; Boontem, P.; Reiter, R.J.; Satayavivad, J. Mechanisms of Melatonin in Alleviating Alzheimer’s Disease. Curr. Neuropharmacol. 2017, 15, 1010–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skarlis, C.; Anagnostouli, M. The role of melatonin in Multiple Sclerosis. Neurol. Sci. 2020, 41, 769–781. [Google Scholar] [CrossRef]
- Zhang, Y.; Cook, A.; Kim, J.; Baranov, S.V.; Jiang, J.; Smith, K.; Cormier, K.; Bennett, E.; Browser, R.P.; Day, A.L.; et al. Melatonin inhibits the caspase-1/cytochrome c/caspase-3 cell death pathway, inhibits MT1 receptor loss and delays disease progression in a mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2013, 55, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Brit. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Laino, D.; D’Alonzo, R.; Mencarelli, A.; di Genova, L.; Fattorusso, A.; Mencaroni, E. Pediatric sleep disturbances and treatment with melatonin. J. Transl. Med. 2019, 17, 77. [Google Scholar] [CrossRef]
- Palumbo, M.L.; Moroni, A.D.; Quiroga, S.; Castro, M.M.; Burgueño, A.L.; Genaro, A.M. Immunomodulation induced by central nervous system-related peptides as a therapeutic strategy for neurodegenerative disorders. Pharmacol. Res. Perspect. 2021, 9, e00795. [Google Scholar] [CrossRef] [PubMed]
- Cicchese, J.M.; Evans, S.; Hult, C.; Joslyn, L.R.; Wessler, T.; Millar, J.A.; Mattila, J.T.; Linderman, J.J.; Kirschner, D.E. Dynamic balance of pro- and anti-inflammatory signals controls disease and limits pathology. Immunol. Rev. 2018, 285, 147–167. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.; Rahu, N. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us? Oxid. Med. Cell. Longev. 2016, 2016, 7432797. [Google Scholar]
- Reiter, R.J.; Mayo, J.C.; Tan, D.X.; Sainz, R.M.; Alatorre-Jimenez, M.; Qin, L.L. Melatonin as an antioxidant: Under promises but over delivers. J. Pineal Res. 2016, 61, 253–278. [Google Scholar] [CrossRef] [PubMed]
- Esposito, E.; Paterniti, I.; Mazzon, E.; Bramanti, P.; Cuzzocrea, S. Melatonin reduces hyperalgesia associated with inflammation. J. Pineal Res. 2010, 49, 321–331. [Google Scholar]
- Hardeland, R. Aging, Melatonin, and the Pro- and Anti-Inflammatory Networks. Int. J. Mol. Sci. 2019, 20, 1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.M.; Kubota, H.; Okita, M.; Maeda, T. The anti-inflammatory and antioxidant effects of melatonin on LPS-stimulated bovine mammary epithelial cells. PLoS ONE 2017, 12, e0178525. [Google Scholar] [CrossRef]
- Tan, D.X.; Manchester, L.C.; Esteban-Zubero, E.; Zhou, Z.; Reiter, R.J. Melatonin as a Potent and Inducible Endogenous Antioxidant: Synthesis and Metabolism. Molecules 2015, 20, 18886–18906. [Google Scholar] [CrossRef] [Green Version]
- Tao, J.; Yang, M.; Wu, H.; Ma, T.; He, C.; Chai, M.; Zhang, X.; Zhang, J.; Ding, F.; Wang, S.; et al. Effects of AANAT overexpression on the inflammatory responses and autophagy activity in the cellular and transgenic animal levels. Autophagy 2018, 14, 1850–1869. [Google Scholar] [CrossRef] [Green Version]
- Rath, M.F.; Coon, S.L.; Amaral, F.G.; Weller, J.L.; Moller, M.; Klein, D.C. Melatonin Synthesis: Acetylserotonin O-Methyltransferase (ASMT) Is Strongly Expressed in a Subpopulation of Pinealocytes in the Male Rat Pineal Gland. Endocrinology 2016, 157, 2028–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talarowska, M.; Szemraj, J.; Zajaczkowska, M.; Galecki, P. ASMT gene expression correlates with cognitive impairment in patients with recurrent depressive disorder. Med. Sci. Monit. 2014, 20, 905–912. [Google Scholar] [PubMed] [Green Version]
- Hu, W.; Tang, J.; Zhang, Z.; Tang, Q.; Yan, Y.; Wang, P.; Wang, X.; Liu, Q.; Guo, X.; Jin, M.; et al. Polymorphisms in the ASMT and ADAMTS1 gene may increase litter size in goats. Vet. Med. Sci. 2020, 6, 775–787. [Google Scholar] [CrossRef]
- Van Leeuwen, S.S.; Te Poele, E.M.; Chatziioannou, A.C.; Benjamins, E.; Haandrikman, A.; Dijkhuizen, L. Goat Milk Oligosaccharides: Their Diversity, Quantity, and Functional Properties in Comparison to Human Milk Oligosaccharides. J. Agric. Food Chem. 2020, 68, 13469–13485. [Google Scholar] [CrossRef] [PubMed]
- Adiguzel, C.; Iqbal, O.; Demir, M.; Fareed, J. European community and US-FDA approval of recombinant human antithrombin produced in genetically altered goats. Clin. Appl. Thromb. Hemost. 2009, 15, 645–651. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.W.; Zhu, H.B.; Du, W.H. Advances of transgenic breeding in livestock. Yi Chuan 2011, 33, 459–468. [Google Scholar] [CrossRef]
- Ma, T.; Tao, J.; Yang, M.; He, C.; Tian, X.; Zhang, X.; Zhang, J.; Deng, S.; Feng, J.; Zhang, Z.; et al. An AANAT/ASMT transgenic animal model constructed with CRISPR/Cas9 system serving as the mammary gland bioreactor to produce melatonin-enriched milk in sheep. J. Pineal Res. 2017, 63, e12406. [Google Scholar] [CrossRef]
- Deng, S.; Wu, Q.; Yu, K.; Zhang, Y.; Yao, Y.; Li, W.; Deng, Z.; Liu, G.; Li, W.; Lian, Z. Changes in the Relative Inflammatory Responses in Sheep Cells Overexpressing of Toll-Like Receptor 4 When Stimulated with LPS. PLoS ONE 2012, 7, e47118. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.D.; Maga, E.A. Genetically engineered livestock for agriculture: A generation after the first transgenic animal research conference. Transgenic Res. 2016, 25, 321–327. [Google Scholar] [CrossRef]
- Robl, J.M.; Wang, Z.; Kasinathan, P.; Kuroiwa, Y. Transgenic animal production and animal biotechnology. Theriogenology 2007, 67, 127–133. [Google Scholar] [CrossRef]
- Bertolini, L.R.; Meade, H.; Lazzarotto, C.R.; Martins, L.T.; Tavares, K.C.; Bertolini, M.; Murray, J.D. The transgenic animal platform for biopharmaceutical production. Transgenic Res. 2016, 25, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Pal, N.; Kertai, M.D.; Lakshminarasimhachar, A.; Avidan, M.S. Pharmacology and clinical applications of human recombinant antithrombin. Expert Opin. Biol. Ther. 2010, 10, 1155–1168. [Google Scholar] [CrossRef]
- Gunata, M.; Parlakpinar, H.; Acet, H.A. Melatonin: A review of its potential functions and effects on neurological diseases. Rev. Neurol. (Paris) 2020, 176, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Pagan, C.; Botros, H.G.; Poirier, K.; Dumaine, A.; Jamain, S.; Moreno, S.; de Brouwer, A.; van Esch, H.; Delorme, R.; Launay, J.M.; et al. Mutation screening of ASMT, the last enzyme of the melatonin pathway, in a large sample of patients with intellectual disability. BMC Med. Genet. 2011, 12, 17. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.; Mora Garcia, M.B. A 100-Year Review: Advances in goat milk research. J. Dairy Sci. 2017, 100, 10026–10044. [Google Scholar] [CrossRef]
- Malik, T.A.; Mohini, M.; Mir, S.H.; Ganaie, B.A.; Singh, D.; Varun, T.K.; Howal, S.; Thakur, S. Somatic Cells in Relation to Udder Health and Milk Quality-A Review. J. Anim. Health Prod. 2018, 6, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Shi, J.; Tian, J.; Tao, J.; Chai, M.; Wang, J.; Xu, Z.; Song, Y.; Zhu, K.; Ji, P.; et al. Exogenous melatonin reduces somatic cell count of milk in Holstein cows. Sci. Rep. 2017, 7, 43280. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Guo, W.; Xu, H.; Tang, K.; Zan, L.; Yang, W. Melatonin suppresses milk fat synthesis by inhibiting the mTOR signaling pathway via the MT1 receptor in bovine mammary epithelial cells. J. Pineal Res. 2019, 67, e12593. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Neglia, G.; Campanile, G.; de Marchi, M. Milk somatic cell count and its relationship with milk yield and quality traits in Italian water buffaloes. J. Dairy Sci. 2020, 103, 5485–5494. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhang, J.; Reiter, R.J.; Ma, X. Melatonin mediates mucosal immune cells, microbial metabolism, and rhythm crosstalk: A therapeutic target to reduce intestinal inflammation. Med. Res. Rev. 2020, 40, 606–632. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Han, H.; Ma, J.; Liu, G.; Wu, X.; Huang, X.; Fang, R.; Baba, K.; Bin, P.; et al. Administration of Exogenous Melatonin Improves the Diurnal Rhythms of the Gut Microbiota in Mice Fed a High-Fat Diet. MSystems 2020, 5, e00002-20. [Google Scholar] [CrossRef]
- Mehaisen, G.M.; Eshak, M.G.; El Sabry, M.I.; Abass, A.O. Expression of Inflammatory and Cell Death Program Genes and Comet DNA Damage Assay Induced by Escherichia coli in Layer Hens. PLoS ONE 2016, 11, e0158314. [Google Scholar] [CrossRef] [Green Version]
- Soares, D.M.; Figueiredo, M.J.; Martins, J.M.; Machado, R.R.; Sorgi, C.; Faciolli, L.H.; Alves-Filho, J.C.; Cunha, F.Q.; Souza, G.E. A crucial role for IL-6 in the CNS of rats during fever induced by the injection of live E. coli. Med. Microbiol. Immunol. 2012, 201, 47–60. [Google Scholar] [CrossRef]
- Sonnweber, T.; Pizzini, A.; Nairz, M.; Weiss, G.; Tancevski, I. Arachidonic Acid Metabolites in Cardiovascular and Metabolic Diseases. Int. J. Mol. Sci. 2018, 23, 3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeds, M.C.; Bass, D.A. Regulation and metabolism of arachidonic acid. Clin. Rev. Allergy Immunol. 1999, 17, 5–26. [Google Scholar] [CrossRef]
- Tilley, S.L.; Coffman, T.M.; Koller, B.H. Mixed messages: Modulation of inflammation and immune responses by prostaglandins and thromboxanes. J. Clin. Investig. 2001, 108, 15–23. [Google Scholar] [CrossRef]
- Fernando, M.R.; Giembycz, M.A.; Mckay, D.M. Bidirectional crosstalk via IL-6, PGE2 and PGD2 between murine myofibroblasts and alternatively activated macrophages enhances anti-inflammatory phenotype in both cells. Br. J. Pharmacol. 2016, 173, 899–912. [Google Scholar] [CrossRef] [Green Version]
- Martinuzzo, M.; del Zar, M.M.; Cardinali, D.P.; Carreras, L.O.; Vacas, M.I. Melatonin effect on arachidonic acid metabolism to cyclooxygenase derivatives in human platelets. J. Pineal Res. 1991, 11, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Stenson, W.F.; Parker, C.W. Metabolism of arachidonic acid in ionophore-stimulated neutrophils. Esterification of a hydroxylated metabolite into phospholipids. J. Clin. Investig. 1979, 64, 1457–1465. [Google Scholar] [CrossRef]
- Tan, D.X.; Hardeland, R. Targeting Host Defense System and Rescuing Compromised Mitochondria to Increase Tolerance against Pathogens by Melatonin May Impact Outcome of Deadly Virus Infection Pertinent to COVID-19. Molecules 2020, 25, 4410. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Xie, Z.; Pei, J.; Wang, B.; Gao, Y.; Qu, Y. Puerarin alters the function of monocytes/macrophages and exhibits chondroprotection in mice. Mol. Med. Rep. 2019, 19, 2876–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Primers Sequence | Tm/°C | Length/bp |
|---|---|---|
| F 5′-ATGTCGGGACATCGTCTTTG-3′ | 58 | 507 |
| R 5′-CATCAGAAGTTAAACAGCACAGTTAG-3′ | 58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Cui, X.; Guan, S.; Li, G.; Yao, Y.; Wu, H.; Zhang, J.; Zhang, X.; Yu, T.; Li, Y.; et al. The Improved Milk Quality and Enhanced Anti-Inflammatory Effect in Acetylserotonin-O-methyltransferase (ASMT) Overexpressed Goats: An Association with the Elevated Endogenous Melatonin Production. Molecules 2022, 27, 572. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020572
Wu H, Cui X, Guan S, Li G, Yao Y, Wu H, Zhang J, Zhang X, Yu T, Li Y, et al. The Improved Milk Quality and Enhanced Anti-Inflammatory Effect in Acetylserotonin-O-methyltransferase (ASMT) Overexpressed Goats: An Association with the Elevated Endogenous Melatonin Production. Molecules. 2022; 27(2):572. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020572
Chicago/Turabian StyleWu, Hao, Xudai Cui, Shengyu Guan, Guangdong Li, Yujun Yao, Haixin Wu, Jinlong Zhang, Xiaosheng Zhang, Tuan Yu, Yunxiang Li, and et al. 2022. "The Improved Milk Quality and Enhanced Anti-Inflammatory Effect in Acetylserotonin-O-methyltransferase (ASMT) Overexpressed Goats: An Association with the Elevated Endogenous Melatonin Production" Molecules 27, no. 2: 572. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020572