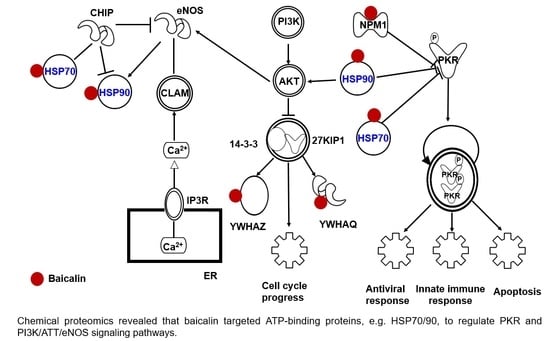
Baicalin Targets HSP70/90 to Regulate PKR/PI3K/AKT/eNOS Signaling Pathways
Abstract
:1. Introduction
2. Results and Discussion
2.1. Assembling and Characterization of Baicalin Functionalized Magnetic Nanoparticles
2.2. Evaluation of BCL-N3 Activity
2.3. The Optimization of Capture of Target Proteins by BCL-N3@MNPs
2.4. Target Proteins of Baicalin
3. Materials and Methods
3.1. Reagents and Materials
3.2. Instruments
3.3. Synthesis of Azido Modified Baicalin Derivative BCL-N3
3.4. Preparation of Baicalin Functionalized Magnetic Nanoparticles BCL-N3@MNP
3.5. Measurement of Biological Activity of Azido-Modified Baicalin
3.6. Cell Culturing and Protein Extraction
3.7. Quantitative Mass Spectrometry Analysis
3.8. Bioinformatics Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gour, A.; Manhas, D.; Bag, S.; Gorain, B.; Nandi, U. Flavonoids as potential phytotherapeutics to combat cytokine storm in SARS-CoV-2. Phytother. Res. 2021, 35, 1–26. [Google Scholar] [CrossRef]
- Banik, K.; Khatoon, E.; Harsha, C.; Rana, V.; Parama, D.; Thakur, K.K.; Bishayee, A.; Kunnumakkara, A.B. Wogonin and its analogs for the prevention and treatment of cancer: A systematic review. Phytother. Res. 2022, 1–30. [Google Scholar] [CrossRef]
- Diaz, P.; Jeong, S.C.; Lee, S.; Khoo, C.; Koyyalamudi, S.R. Antioxidant and anti-inflammatory activities of selected medicinal plants and fungi containing phenolic and flavonoid compounds. Chin. Med. 2012, 7, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Li, B.Q.; Fu, T.; Gong, W.H.; Dunlop, N.; Kung, H.F.; Yan, Y.D.; Kang, J.; Wang, J.M. The flavonoid baicalin exhibits anti-inflammatory activity by binding to chemokines. Immunopharmacology 2000, 49, 295–306. [Google Scholar] [CrossRef]
- Picciolo, G.; Mannino, F.; Irrera, N.; Minutoli, L.; Altavilla, D.; Vaccaro, M.; Oteri, G.; Squadrito, F.; Pallio, G. Reduction of oxidative stress blunts the NLRP3 inflammatory cascade in LPS stimulated human gingival fibroblasts and oral mucosal epithelial cells. Biomed. Pharmacother. 2022, 146, 112525. [Google Scholar] [CrossRef]
- Xu, G.; Dou, J.; Zhang, L.; Guo, Q.; Zhou, C. Inhibitory Effects of Baicalein on the Influenza Virus in Vivo Is Determined by Baicalin in the Serum. Biol. Pharm. Bull. 2010, 33, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Tsou, L.K.; Lara-Tejero, M.; RoseFigura, J.; Zhang, Z.J.; Wang, Y.-C.; Yount, J.S.; Lefebre, M.; Dossa, P.D.; Kato, J.; Guan, F.; et al. Antibacterial Flavonoids from Medicinal Plants Covalently Inactivate Type III Protein Secretion Substrates. J. Am. Chem. Soc. 2016, 138, 2209–2218. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, K.; Honda, M.; Yoshizaki, H.; Yamamoto, S.; Nakane, H.; Fukushima, M.; Ono, K.; Tokunaga, T. Baicalin, an inhibitor of HIV-1 production in vitro. Antivir. Res. 1998, 37, 131–140. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Lin, M.-T.; Wang, J.-J.; Liao, J.-F.; Huang, W.-T. The antipyretic effects of baicalin in lipopolysaccharide-evoked fever in rabbits. Neuropharmacology 2006, 51, 709–717. [Google Scholar] [CrossRef]
- Chan, F.L.; Choi, H.L.; Chen, Z.Y.; Chan, P.S.F.; Huang, Y. Induction of apoptosis in prostate cancer cell lines by a flavonoid, baicalin. Cancer Lett. 2000, 160, 219–228. [Google Scholar] [CrossRef]
- Dai, J.; Liang, K.; Zhao, S.; Jia, W.; Liu, Y.; Wu, H.; Lv, J.; Cao, C.; Chen, T.; Zhuang, S.; et al. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc. Natl. Acad. Sci. USA 2018, 115, E5896–E5905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Toume, K.; Luvai, E.; Nwe, K.M.; Mizukami, S.; Hirayama, K.; Komatsu, K.; Morita, K. The discovery of herbal drugs and natural compounds as inhibitors of SARS-CoV-2 infection in vitro. J. Nat. Med. 2022, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yao, Q.; An, Y.; Fan, L.; Wang, J.; Li, H. Baicalin suppresses the progression of Type 2 diabetes-induced liver tumor through regulating METTL3/m6A/HKDC1 axis and downstream p-JAK2/STAT1/clevaged Capase3 pathway. Phytomedicine 2022, 94, 153823. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, S.; Cheng, Y.; Lu, Y.; Jia, Z.; Yang, X.; Zhang, S.; Guo, W.; Pei, L. Baicalin suppresses neuron autophagy and apoptosis by regulating astrocyte polarization in pentylenetetrazol-induced epileptic rats and PC12 cells. Brain Res. 2022, 1774, 147723. [Google Scholar] [CrossRef]
- Sahebkar, A. Baicalin as a potentially promising drug for the management of sulfur mustard induced cutaneous complications: A review of molecular mechanisms. Cutan. Ocul. Toxicol. 2012, 31, 226–234. [Google Scholar] [CrossRef]
- Xin, W.; Song, J.; He, G.; Du, G. Progress in pharmacological study and the underlying mechanism of baicalein and baicalin. Chin. J. New Drugs. 2013, 22, 647–653. [Google Scholar]
- Zhu, Y.; Yang, Q.; Zhang, S.; Zhang, M.; Gao, X. Advances in pharmacological effects and mechanisms of baicalin and baicalein. Lishizhen Med. Mater. Med. Res. 2020, 31, 921–925. [Google Scholar]
- Anwar, S.; Mohammad, T.; Shamsi, A.; Queen, A.; Parveen, S.; Luqman, S.; Hasan, G.M.; Alamry, K.A.; Azum, N.; Asiri, A.M.; et al. Discovery of Hordenine as a Potential Inhibitor of Pyruvate Dehydrogenase Kinase 3: Implication in Lung Cancer Therapy. Biomedicines 2020, 8, 119. [Google Scholar] [CrossRef]
- Laggner, C.; Kokel, D.; Setola, V.; Tolia, A.; Lin, H.; Irwin, J.J.; Keiser, M.J.; Cheung, C.Y.J.; Minor, D.L., Jr.; Roth, B.L.; et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat. Chem. Biol. 2012, 8, 144–146. [Google Scholar] [CrossRef]
- Sato, S.-i.; Kwon, Y.; Kamisuki, S.; Srivastava, N.; Mao, Q.; Kawazoe, Y.; Uesugi, M. Polyproline-rod approach to isolating protein targets of bioactive small molecules: Isolation of a new target of indomethacin. J. Am. Chem. Soc. 2007, 129, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Mohammad, T.; Anwar, S.; AlAjmi, M.F.; Hussain, A.; Rehman, M.; Islam, A.; Hassan, M. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: Possible implication in COVID-19 therapy. Biosci. Rep. 2020, 40, BSR2020125623. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, S.; Anjum, H.; Shahbaaz, M.; Khan, M.S.; Ataya, F.S.; Alamri, A.; Alhumaydhi, F.A.; Husain, F.M.; Rehman, M.T.; Mohammad, T.; et al. A computational study on active constituents of Habb-ul-aas and Tabasheer as inhibitors of SARS-CoV-2 main protease. J. Biomol. Struct. Dyn. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bantscheff, M.; Scholten, A.; Heck, A.J.R. Revealing promiscuous drug-target interactions by chemical proteomics. Drug Discov. Today. 2009, 14, 1021–1029. [Google Scholar] [CrossRef]
- Cheung, A.K.; Jain, R.K. Accelerating the discovery of new drug targets with chemical proteomics. Idrugs 2010, 13, 862–868. [Google Scholar]
- Rix, U.; Superti-Furga, G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009, 5, 616–624. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Ma, N.; Tian, J.; Shao, Y.; Zhu, B.; Wong, Y.K.; Liang, Z.; Zou, C.; Wang, J. Target identification of natural medicine with chemical proteomics approach: Probe synthesis, target fishing and protein identification. Signal. Transduct. Target. Ther. 2020, 5, 72. [Google Scholar] [CrossRef]
- Barglow, K.T.; Cravatt, B.F. Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods 2007, 4, 822–827. [Google Scholar] [CrossRef]
- Katayama, H.; Oda, Y. Chemical proteomics for drug discovery based on compound-immobilized affinity chromatography. J. Chromatogr. B 2007, 855, 21–27. [Google Scholar] [CrossRef]
- Kanoh, N.; Honda, K.; Simizu, S.; Muroi, M.; Osada, H. Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew. Chem. Int. Ed. 2005, 44, 3559–3562. [Google Scholar] [CrossRef]
- Sakamoto, S.; Kabe, Y.; Hatakeyama, M.; Yamaguchi, Y.; Handa, H. Development and Application of High-Performance Affinity Beads: Toward Chemical Biology and Drug Discovery. Chem. Rec. 2009, 9, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Luo, Q.; Yang, L.; Bing, T.; Li, X.; Guo, W.; Wu, K.; Zhao, Y.; Xiong, S.; Shangguan, D.; et al. Mass Spectrometric Proteomics Reveals that Nuclear Protein Positive Cofactor PC4 Selectively Binds to Cross-Linked DNA by a trans-Platinum Anticancer Complex. J. Am. Chem. Soc. 2014, 136, 2948–2951. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Hou, Y.; Hou, J.; Pan, P.; Li, L.-Y.; Bai, G.; Luo, G. Preparation of Functionalized Alkynyl Magnetic Microspheres for the Selective Enrichment of Cell Glycoproteins Based on Click Chemistry. Biomacromolecules 2013, 14, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.-M.; Yu, J.; Kim, H.; Lee, N.-R.; Kim, S.W.; Lee, N.-J.; Lee, J.; Seong, J.; Kim, N.-J.; Inn, K.-S. Identification of actin as a direct proteomic target of berberine using an affinity-based chemical probe and elucidation of its modulatory role in actin assembly. Chem. Commun. 2017, 53, 7045–7047. [Google Scholar] [CrossRef]
- Adalsteinsson, O.; Lamotte, A.; Baddour, R.F.; Colton, C.K.; Pollak, A.; Whitesides, G.M. Preparation and magnetic filtration of polyacrylamide gels containing covalently immobilized proteins and a ferrofluid. J. Mol. Catal. 1979, 6, 199–225. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Deng, C. Functionalized magnetic nanoparticles for sample preparation in proteomics and peptidomics analysis. Chem. Soc. Rev. 2013, 42, 8517–8539. [Google Scholar] [CrossRef]
- Grimsdale, A.C.; Mullen, K. The chemistry of organic nanomaterials. Angew. Chem. Int. Ed. 2005, 44, 5592–5629. [Google Scholar] [CrossRef]
- Frey, N.A.; Peng, S.; Cheng, K.; Sun, S. Magnetic nanoparticles: Synthesis, functionalization, and applications in bioimaging and magnetic energy storage. Chem. Soc. Rev. 2009, 38, 2532–2542. [Google Scholar] [CrossRef]
- Zeng, W.J.; Du, Z.F.; Zhao, Q.; Zhao, Y.; Wang, Y.Y.; Wu, K.; Jia, F.F.; Zhang, Y.Y.; Wang, F.Y. Proteomic Strategy for Identification of Proteins Responding to Cisplatin-Damaged DNA. Anal. Chem. 2019, 91, 6035–6042. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, W.; Xiao, Y. Profiling of Multiple Targets of Artemisinin Activated by Hemin in Cancer Cell Proteome. ACS Chem. Biol. 2016, 11, 882–888. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.J.; Chia, W.N.; Loh, C.C.; Li, Z.; Lee, Y.M.; He, Y.; Yuan, L.X.; Lim, T.K.; Liu, M. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 2015, 6, 10111–10121. [Google Scholar] [CrossRef]
- Ismail, H.M.; Barton, V.; Phanchana, M.; Charoensutthivarakul, S.; Wong, M.H.; Hemingway, J.; Biagini, G.A.; O’Neill, P.M.; Ward, S.A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. USA 2016, 113, 2080–2085. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhou, G.B.; Liu, P.; Song, J.H.; Liang, Y.; Yan, X.J.; Xu, F.; Wang, B.S.; Mao, J.H.; Shen, Z.X. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 4826–4831. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.W.; Yan, X.J.; Zhou, Z.R.; Yang, F.F.; Wu, Z.Y.; Sun, H.B.; Liang, W.X.; Song, A.X.; Lallemand-Breitenbach, V.; Jeanne, M. Arsenic Trioxide Controls the Fate of the PML-RARα Oncoprotein by Directly Binding PML. Science 2010, 328, 240–243. [Google Scholar] [CrossRef]
- Wang, D.; Cao, Y.; Zheng, L.; Lv, D.; Chen, L.; Xing, X.; Zhu, Z.; Li, X.; Chai, Y. Identification of Annexin A2 as a target protein for plant alkaloid matrine. Chem. Commun. 2017, 53, 5020–5023. [Google Scholar] [CrossRef]
- Wang, D.-D.; Jin, Q.; Zou, L.-W.; Hou, J.; Lv, X.; Lei, W.; Cheng, H.-L.; Ge, G.-B.; Yang, L. A bioluminescent sensor for highly selective and sensitive detection of human carboxylesterase 1 in complex biological samples. Chem. Commun. 2016, 52, 3183–3186. [Google Scholar] [CrossRef]
- Bong, S.M.; Moon, J.H.; Nam, K.H.; Lee, K.S.; Chi, Y.M.; Hwang, K.Y. Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3--creatine transition-state analogue complex. Febs Lett 2008, 582, 3959–3965. [Google Scholar] [CrossRef] [Green Version]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperon 2016, 21, 379–404. [Google Scholar] [CrossRef] [Green Version]
- Pearl, L.H. The HSP90 Molecular Chaperone-An Enigmatic ATPase. Biopolymers 2016, 105, 594–607. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Goyal, S.; Jamal, S.; Singh, A.; Grover, A. Hsp90: Friends, clients and natural foes. Biochimie 2016, 127, 227–240. [Google Scholar] [CrossRef]
- Kaitsuka, T.; Tomizawa, K.; Matsushita, M. Transformation of eEF1B delta into heat-shock response transcription factor by alternative splicing. Embo Rep. 2011, 12, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Fujita, N.; Tsuruo, T. Regulation of kinase activity of 3-phosphoinositide-dependent protein kinase-1 by binding to 14-3-3. J. Biol. Chem. 2002, 277, 39360–39367. [Google Scholar] [CrossRef] [Green Version]
- Popov, I.K.; Hiatt, S.M.; Whalen, S.; Keren, B.; Ruivenkamp, C.; van Haeringen, A.; Chen, M.J.; Cooper, G.M.; Korf, B.R.; Chang, C.B. A YWHAZ Variant Associated With Cardiofaciocutaneous Syndrome Activates the RAF-ERK Pathway. Front. Physiol. 2019, 10, 388. [Google Scholar] [CrossRef] [Green Version]
- Herwald, H.; Dedio, J.; Kellner, R.; Loos, M.; MullerEsterl, W. Isolation and characterization of the kininogen-binding protein p33 from endothelial cells - Identity with the gC1q receptor. J. Biol. Chem. 1996, 271, 13040–13047. [Google Scholar] [CrossRef] [Green Version]
- Joseph, K.; Ghebrehiwet, B.; Peerschke, E.I.B.; Reid, K.B.M.; Kaplan, A.P. Identification of the zinc-dependent endothelial cell binding protein for high molecular weight kininogen and factor XII: Identity with the receptor that binds to the globular “heads” of C1q (gC1q-R). Proc. Natl. Acad. Sci. USA 1996, 93, 8552–8557. [Google Scholar] [CrossRef] [Green Version]
- Leigh, L.E.A.; Ghebrehiwet, B.; Perera, T.P.S.; Bird, I.N.; Strong, P.; Kishore, U.; Reid, K.B.M.; Eggleton, P. Clq-mediated chemotaxis by human neutrophils: Involvement of gClqR and G-protein signalling mechanisms. Biochem. J. 1998, 330, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Feichtinger, R.G.; Olahova, M.; Kishita, Y.; Garone, C.; Kremer, L.S.; Yagi, M.; Uchiumi, T.; Jourdain, A.A.; Thompson, K.; D’Souza, A.R.; et al. Biallelic C1QBP Mutations Cause Severe Neonatal-, Childhood-, or Later-Onset Cardiomyopathy Associated with Combined Respiratory-Chain Deficiencies. Am. J. Hum. Genet. 2017, 101, 525–538. [Google Scholar] [CrossRef]
- Hosszu, K.K.; Valentino, A.; Vinayagasundaram, U.; Vinayagasundaram, R.; Joyce, M.G.; Ji, Y.; Peerschke, E.I.B.; Ghebrehiwet, B. DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells. Blood 2012, 120, 1228–1236. [Google Scholar] [CrossRef] [Green Version]
- Pixley, R.A.; Espinola, R.G.; Ghebrehiwet, B.; Joseph, K.; Kao, A.; Bdeir, K.; Cines, D.B.; Colman, R.W. Interaction of high-molecular-weight kininogen with endothelial cell binding proteins suPAR, gC1qR and cytokeratin 1 determined by Surface Plasmon Resonance (BiaCore). Thromb. Haemost. 2011, 105, 1053–1059. [Google Scholar]
- Yoshikawa, H.; Komatsu, W.; Hayano, T.; Miura, Y.; Homma, K.; Izumikawa, K.; Ishikawa, H.; Miyazawa, N.; Tachikawa, H.; Yamauchi, Y.; et al. Splicing Factor 2-Associated Protein p32 Participates in Ribosome Biogenesis by Regulating the Binding of Nop52 and Fibrillarin to Preribosome Particles. Mol. Cell. Proteom. 2011, 10, M110.006148. [Google Scholar] [CrossRef] [Green Version]
- Swaminathan, V.; Kishore, A.H.; Febitha, K.K.; Kundu, T.K. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 2005, 25, 7534–7545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, Y.; Park, B.; Koh, W.; Lee, S.; Cheon, Y.; Kim, R.; Che, L.; Lee, S. New Centromeric Component CENP-W Is an RNA-associated Nuclear Matrix Protein That Interacts with Nucleophosmin/B23 Protein. J. Biol. Chem. 2011, 286, 42758–42769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Cho, Y.E.; Park, J.H. The Nucleolar Protein GLTSCR2 Is an Upstream Negative Regulator of the Oncogenic Nucleophosmin-MYC Axis. Am. J. Pathol. 2015, 185, 2061–2068. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.T.; Alekseev, O.M.; Grossman, G.; Widgren, E.E.; Thresher, R.; Wagner, E.J.; Sullivan, K.D.; Marzluff, W.F.; O’Rand, M.G. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 2006, 281, 21526–21534. [Google Scholar] [CrossRef] [Green Version]
- Jin, D.Y.; Chae, H.Z.; Rhee, S.G.; Jeang, K.T. Regulatory role for a novel human thioredoxin peroxidase in NF-kappa B activation. J. Biol. Chem. 1997, 272, 30952–30961. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Yamada, T.; Endo, R.; Ino, Y.; Gotoh, M.; Tsuda, H.; Yamada, Y.; Chiba, H.; Hirohashi, S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J. Cell. Biol. 1998, 140, 1383–1393, Correction in J. Cell. Biol. 1998, 143, 277–277. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.K.; Wang, X.; Sun, F.; Wang, C.C. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem. J. 2012, 441, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; Nakamura, T.; Inouye, K.; Li, J.H.; Tang, Y.T.; Lundback, P.; Valdes-Ferrer, S.I.; Olofsson, P.S.; Kalb, T.; Roth, J.; et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 2012, 488, 670–674. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Franchi, L.; Nunez, G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur. J. Immunol. 2013, 43, 1147–1152. [Google Scholar] [CrossRef] [Green Version]
- Engelman, J.A. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat. Rev. Cancer. 2009, 9, 550–562. [Google Scholar] [CrossRef]
- Anwar, S.; Shamsi, A.; Mohammad, T.; Islam, A.; Hassan, M.I. Targeting pyruvate dehydrogenase kinase signaling in the development of effective cancer therapy. Biochim. Biophys. Acta Rev. Cancer. 2021, 1876, 188568. [Google Scholar] [CrossRef] [PubMed]
- Sessa, W.C. eNOS at a glance. J. Cell. Sci. 2004, 117, 2427–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malekmohammad, K.; Sewell, R.D.E.; Rafieian-Kopaei, M. Mechanisms of Medicinal Plant Activity on Nitric Oxide (NO) Bioavailability as Prospective Treatments for Atherosclerosis. Curr. Pharm. Des. 2020, 26, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]








| Gene Name | Protein Name (Abbreviation) | Protein Type | GO Information | |
|---|---|---|---|---|
| Biological Process | Molecular Function | |||
| CKB | Creatine kinase B-type (CKB) | kinase | Creatine metabolism process, phosphocreatine biosynthesis | ATP binding, creatine kinase activity |
| YWHAQ | 14-3-3 protein theta (14-3-3η) | enzyme | Negative regulation of ion transmembrane transport and transcription | 14-3-3 protein binding, ion channel binding |
| C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial (C1qBP) | transcription regulator | Adaptive immunity response, host-virusC1qBP interaction, mRNA splicing | Adrenergic receptor binding, complement component C1q complex binding |
| YWHAZ | 14-3-3 protein zeta/delta (14-3-3φ/δ) | other | Adaptive immune response, cytokine-mediated signaling, Golgi reassembly | Cadherin binding, ion channel binding, protein kinase binding |
| HSP90AA1 | Heat shock protein HSP 90-alpha (HSP86) | enzyme | Axon extension, response to heat, response to virus, chaperone mediated autophagy | ATPase activity, ATP binding, GTPase binding |
| EEF1D | Elongation factor 1-delta (EF-1-δ) | translation regulator | Cellular response to ionizing radiation, mRNA transcription | Activating transcription factor binding, cadherin binding, DNA binding |
| HSPA1A | Heat shock 70 kDa protein 1A, (HSP70-1) | enzyme | ATP metabolism, cellular heat acclimation, response to oxidative stress | ATPase activity, ATP binding, cadherin binding |
| HSP90AB1 | Heat shock protein HSP 90-beta (HSP 90) | enzyme | Axon extension, response to heat, response to interleukin-4 | ATPase activity, ATP binding, ATP-dependent protein binding |
| NPM1 | Nucleophosmin (NPM) | transcription regulator | Cell aging, centrosome cycle, DNA repair, intracellular protein transport | Activating transcription factor binding, chromatin binding, RNA binding |
| ATP5F1B | ATP synthase subunit beta, (ATPSF1β) | transporter | ATP biosynthesis, lipid metabolism, mitochondrion organization | ATP binding, ATPase activity |
| ACTG1 | Actin, cytoplasmic 2 (ACTP2) | other | Angiogenesis, positive regulation of cell migration, retina homeostasis | ATP binding, profiling binding |
| ACTN4 | Alpha-actinin-4 (ACTN4) | transcription regulator | Protein transport, positive regulation of cell migration, platelet degranulation | Actin binding, calcium ion binding, chromatin DNA binding |
| NASP | Nuclear autoantigenic sperm protein (NASP) | other | Nucleosome assembly, DNA replication, histone exchange | Histone binding |
| PRDX4 | Peroxiredoxin-4 (Prx-IV) | enzyme | Cell redox homeostasis, extracellular matrix organization, I-kappaB phosphorylation | Thioredoxin peroxidase activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Liang, Z.; Qi, L.; Tang, C.; Liu, X.; Tang, J.; Zhao, Y.; Zhang, Y.; Fang, T.; Luo, Q.; et al. Baicalin Targets HSP70/90 to Regulate PKR/PI3K/AKT/eNOS Signaling Pathways. Molecules 2022, 27, 1432. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27041432
Hou Y, Liang Z, Qi L, Tang C, Liu X, Tang J, Zhao Y, Zhang Y, Fang T, Luo Q, et al. Baicalin Targets HSP70/90 to Regulate PKR/PI3K/AKT/eNOS Signaling Pathways. Molecules. 2022; 27(4):1432. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27041432
Chicago/Turabian StyleHou, Yinzhu, Zuqing Liang, Luyu Qi, Chao Tang, Xingkai Liu, Jilin Tang, Yao Zhao, Yanyan Zhang, Tiantian Fang, Qun Luo, and et al. 2022. "Baicalin Targets HSP70/90 to Regulate PKR/PI3K/AKT/eNOS Signaling Pathways" Molecules 27, no. 4: 1432. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27041432