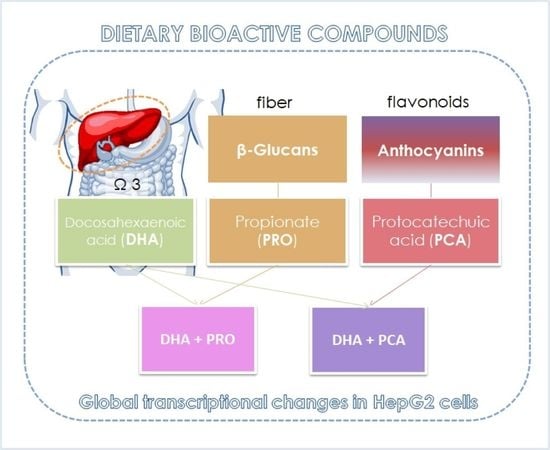
Co-Administration of Propionate or Protocatechuic Acid Does Not Affect DHA-Specific Transcriptional Effects on Lipid Metabolism in Cultured Hepatic Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. HepG2 Cell Culture and Supplementation
2.3. Lipid Extraction and Fatty Acid Composition Analysis
2.4. Isolation of RNA, RNA Sequencing (RNA-seq) and Quantitative Real-Time PCR (RT-qPCR)
2.5. Intracellular Triglyceride (TG) Content and Cholesterol Secretion
2.6. Statistical Analysis
2.6.1. Biochemical and RT-qPCR Data Analysis
2.6.2. RNA-Seq Analysis
3. Results and Discussion
4. Conclusions
Availability of Supporting Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Binesh Marvasti, T.; Adeli, K. Pharmacological management of metabolic syndrome and its lipid complications. Daru 2010, 18, 146–154. [Google Scholar] [PubMed]
- Bordoni, A.; Boesch, C.; Malpuech-Brugère, C.; Orfila, C.; Tomás-Cobos, L. The role of bioactives in energy metabolism and metabolic syndrome. Proc. Nutr. Soc. 2019, 78, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Albracht-Schulte, K.; Kalupahana, N.S.; Ramalingam, L.; Wang, S.; Rahman, S.M.; Robert-McComb, J.; Moustaid-Moussa, N. Omega-3 fatty acids in obesity and metabolic syndrome: A mechanistic update. J. Nutr. Biochem. 2018, 58, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bellenger, J.; Bellenger, S.; Escoula, Q.; Bidu, C.; Narce, M. n-3 Polyunsaturated fatty acids: An innovative strategy against obesity and related metabolic disorders, intestinal alteration and gut microbiota dysbiosis. Biochimie 2019, 159, 66–71. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M.; Richter, C.K.; Bowen, K.J.; Skulas-Ray, A.C.; Jackson, K.H.; Petersen, K.S.; Harris, W.S. Recent clinical trials shed new light on the cardiovascular benefits of omega-3 fatty acids. Methodist Debakey Cardiovasc. J. 2019, 15, 171–178. [Google Scholar]
- O’Mahoney, L.L.; Matu, J.; Price, O.J.; Birch, K.M.; Ajjan, R.A.; Farrar, D.; Tapp, R.; West, D.J.; Deighton, K.; Campbell, M.D. Omega-3 polyunsaturated fatty acids favourably modulate cardiometabolic biomarkers in type 2 diabetes: A meta-analysis and meta-regression of randomized controlled trials. Cardiovasc. Diabetol. 2018, 17, 98. [Google Scholar] [CrossRef]
- Guo, X.F.; Sinclair, A.J.; Kaur, G.; Li, D. Differential effects of EPA, DPA and DHA on cardio-metabolic risk factors in high-fat diet fed mice. Prostaglandins Leukot. Essent. Fatty Acids 2018, 136, 47–55. [Google Scholar] [CrossRef]
- Vas Dias, F.W. 13—Authorised EU health claims for DHA and EPA. In Foods, Nutrients and Food Ingredients with Authorised EU Health Claims; Sadler, M.J., Ed.; Woodhead Publishing: Oxford, UK, 2015; Volume 2, pp. 237–256. [Google Scholar]
- Bordoni, A.; Di Nunzio, M.; Danesi, F.; Biagi, P.L. Polyunsaturated fatty acids: From diet to binding to PPARs and other nuclear receptors. Genes Nutr. 2006, 1, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Soni, N.; Ross, A.B.; Scheers, N.; Nookaew, I.; Gabrielsson, B.G.; Sandberg, A.S. The omega-3 fatty acids EPA and DHA, as a part of a murine high-fat diet, reduced lipid accumulation in brown and white adipose tissues. Int. J. Mol. Sci. 2019, 20, 5895. [Google Scholar] [CrossRef] [Green Version]
- Innes, J.K.; Calder, P.C. The differential effects of eicosapentaenoic acid and docosahexaenoic acid on cardiometabolic risk factors: A systematic review. Int. J. Mol. Sci. 2018, 19, 532. [Google Scholar] [CrossRef] [Green Version]
- Ho, H.V.; Sievenpiper, J.L.; Zurbau, A.; Blanco Mejia, S.; Jovanovski, E.; Au-Yeung, F.; Jenkins, A.L.; Vuksan, V. The effect of oat β-glucan on LDL-cholesterol, non-HDL-cholesterol and apoB for CVD risk reduction: A systematic review and meta-analysis of randomised-controlled trials. Br. J. Nutr. 2016, 116, 1369–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caz, V.; Gil-Ramírez, A.; Largo, C.; Tabernero, M.; Santamaría, M.; Martín-Hernández, R.; Marín, F.R.; Reglero, G.; Soler-Rivas, C. Modulation of cholesterol-related gene expression by dietary fiber fractions from edible mushrooms. J. Agric. Food Chem. 2015, 63, 7371–7380. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, E.; Grootaert, C.; Verstraete, W.; Van de Wiele, T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011, 69, 245–258. [Google Scholar] [CrossRef]
- Reis, J.F.; Monteiro, V.V.; de Souza Gomes, R.; do Carmo, M.M.; da Costa, G.V.; Ribera, P.C.; Monteiro, M.C. Action mechanism and cardiovascular effect of anthocyanins: A systematic review of animal and human studies. J. Transl. Med. 2016, 14, 315. [Google Scholar] [CrossRef] [Green Version]
- Du, C.; Shi, Y.; Ren, Y.; Wu, H.; Yao, F.; Wei, J.; Wu, M.; Hou, Y.; Duan, H. Anthocyanins inhibit high-glucose-induced cholesterol accumulation and inflammation by activating LXRα pathway in HK-2 cells. Drug Des. Dev. Ther. 2015, 9, 5099–5113. [Google Scholar]
- Tsuda, T.; Horio, F.; Uchida, K.; Aoki, H.; Osawa, T. Dietary cyanidin 3-O-β-D-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 2003, 133, 2125–2130. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Valli, V.; Tomas-Cobos, L.; Tomas-Chisbert, T.; Murgui-Bosch, L.; Danesi, F.; Bordoni, A. Is cytotoxicity a determinant of the different in vitro and in vivo effects of bioactives? BMC Complement. Altern. Med. 2017, 17, 453. [Google Scholar] [CrossRef] [Green Version]
- Ghini, V.; Di Nunzio, M.; Tenori, L.; Valli, V.; Danesi, F.; Capozzi, F.; Luchinat, C.; Bordoni, A. Evidence of a DHA signature in the lipidome and metabolome of human hepatocytes. Int. J. Mol. Sci. 2017, 18, 359. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Madsen, J.G.S.; Schmidt, S.F.; Larsen, B.D.; Loft, A.; Nielsen, R.; Mandrup, S. iRNA-seq: Computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 2015, 43, e40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danesi, F.; Kroon, P.A.; Saha, S.; de Biase, D.; D’Antuono, L.F.; Bordoni, A. Mixed pro- and anti-oxidative effects of pomegranate polyphenols in cultured cells. Int. J. Mol. Sci. 2014, 15, 19458–19471. [Google Scholar] [CrossRef] [PubMed]
- Danesi, F.; Govoni, M.; D’Antuono, L.F.; Bordoni, A. The molecular mechanism of the cholesterol-lowering effect of dill and kale: The influence of the food matrix components. Electrophoresis 2016, 37, 1805–1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jump, D.B.; Botolin, D.; Wang, Y.; Xu, J.; Demeure, O.; Christian, B. Docosahexaenoic acid (DHA) and hepatic gene transcription. Chem. Phys. Lipids 2008, 153, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Arai, T.; Kim, H.J.; Chiba, H.; Matsumoto, A. Interaction of fenofibrate and fish oil in relation to lipid metabolism in mice. J. Atheroscler. Thromb. 2009, 16, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Sinclair, A.J.; Cameron-Smith, D.; Barr, D.P.; Molero-Navajas, J.C.; Konstantopoulos, N. Docosapentaenoic acid (22:5n-3) down-regulates the expression of genes involved in fat synthesis in liver cells. Prostaglandins Leukot. Essent. Fatty Acids 2011, 85, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Mazein, A.; Watterson, S.; Hsieh, W.Y.; Griffiths, W.J.; Ghazal, P. A comprehensive machine-readable view of the mammalian cholesterol biosynthesis pathway. Biochem. Pharmacol. 2013, 86, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapani, L.; Segatto, M.; Pallottini, V. Regulation and deregulation of cholesterol homeostasis: The liver as a metabolic “power station”. World J. Hepatol. 2012, 4, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Honda, A.; Salen, G.; Nguyen, L.B.; Tint, G.S.; Batta, A.K.; Shefer, S. Down-regulation of cholesterol biosynthesis in sitosterolemia: Diminished activities of acetoacetyl-CoA thiolase, 3-hydroxy-3-methylglutaryl-CoA synthase, reductase, squalene synthase, and 7-dehydrocholesterol delta7-reductase in liver and mononuclear leukocytes. J. Lipid Res. 1998, 39, 44–50. [Google Scholar] [PubMed]
- Botolin, D.; Wang, Y.; Christian, B.; Jump, D.B. Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. J. Lipid Res. 2006, 47, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimomura, I.; Shimano, H.; Horton, J.D.; Goldstein, J.L.; Brown, M.S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Investig. 1997, 99, 838–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, J.T.; Guryev, O.; Brown, M.S.; Goldstein, J.L. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J. Biol. Chem. 1998, 273, 26138–26148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, G.F.; Rader, D.J. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005, 96, 1221–1232. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, C.; Huang, S.; Gong, B.; Yu, J.; Shi, Q.; Chen, G. Effects of individual and multiple fatty acids (palmitate, oleate and docosahaexenoic acid) on cell viability and lipid metabolism in LO2 human liver cells. Mol. Med. Rep. 2014, 10, 3254–3260. [Google Scholar] [CrossRef] [Green Version]
- Righi, V.; Di Nunzio, M.; Danesi, F.; Schenetti, L.; Mucci, A.; Boschetti, E.; Biagi, P.; Bonora, S.; Tugnoli, V.; Bordoni, A. EPA or DHA supplementation increases triacylglycerol, but not phospholipid, levels in isolated rat cardiomyocytes. Lipids 2011, 46, 627–636. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Valli, V.; Bordoni, A. Pro- and anti-oxidant effects of polyunsaturated fatty acid supplementation in HepG2 cells. Prostaglandins Leukot. Essent. Fatty Acids 2011, 85, 121–127. [Google Scholar] [CrossRef]
- Calder, P.C. Very long-chain n-3 fatty acids and human health: Fact, fiction and the future. Proc. Nutr. Soc. 2018, 77, 52–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Hu, F.B.; Manson, J.E. Marine omega-3 supplementation and cardiovascular disease: An updated meta-analysis of 13 randomized controlled trials involving 127,477 participants. J. Am. Heart Assoc. 2019, 8, e013543. [Google Scholar] [CrossRef] [PubMed]
- Kunz, H.E.; Dasari, S.; Lanza, I.R. EPA and DHA elicit distinct transcriptional responses to high-fat feeding in skeletal muscle and liver. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E460–E472. [Google Scholar] [CrossRef] [PubMed]
- Metherel, A.H.; Irfan, M.; Klingel, S.L.; Mutch, D.M.; Bazinet, R.P. Compound-specific isotope analysis reveals no retroconversion of DHA to EPA but substantial conversion of EPA to DHA following supplementation: A randomized control trial. Am. J. Clin. Nutr. 2019, 110, 823–831. [Google Scholar] [CrossRef]
- Chénais, B.; Cornec, M.; Dumont, S.; Marchand, J.; Blanckaert, V. Transcriptomic response of breast cancer cells MDA-MB-231 to docosahexaenoic acid: Downregulation of lipid and cholesterol metabolism genes and upregulation of genes of the pro-apoptotic ER-stress pathway. Int. J. Environ. Res. Public Health 2020, 17, 3746. [Google Scholar] [CrossRef]
- Bub, A.; Malpuech-Brugère, C.; Orfila, C.; Amat, J.; Arianna, A.; Blot, A.; Di Nunzio, M.; Holmes, M.; Kertész, Z.; Marshall, L.; et al. A dietary intervention of bioactive enriched foods aimed at adults at risk of metabolic syndrome: Protocol and results from PATHWAY-27 pilot study. Nutrients 2019, 11, 1814. [Google Scholar] [CrossRef] [Green Version]
- Ghini, V.; Tenori, L.; Capozzi, F.; Luchinat, C.; Bub, A.; Malpuech-Brugere, C.; Orfila, C.; Ricciardiello, L.; Bordoni, A. DHA-induced perturbation of human serum metabolome. Role of the food matrix and co-administration of oat β-glucan and anthocyanins. Nutrients 2019, 12, 86. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danesi, F.; Larsen, B.D.; Di Nunzio, M.; Nielsen, R.; de Biase, D.; Valli, V.; Mandrup, S.; Bordoni, A. Co-Administration of Propionate or Protocatechuic Acid Does Not Affect DHA-Specific Transcriptional Effects on Lipid Metabolism in Cultured Hepatic Cells. Nutrients 2020, 12, 2952. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12102952
Danesi F, Larsen BD, Di Nunzio M, Nielsen R, de Biase D, Valli V, Mandrup S, Bordoni A. Co-Administration of Propionate or Protocatechuic Acid Does Not Affect DHA-Specific Transcriptional Effects on Lipid Metabolism in Cultured Hepatic Cells. Nutrients. 2020; 12(10):2952. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12102952
Chicago/Turabian StyleDanesi, Francesca, Bjørk D. Larsen, Mattia Di Nunzio, Ronni Nielsen, Dario de Biase, Veronica Valli, Susanne Mandrup, and Alessandra Bordoni. 2020. "Co-Administration of Propionate or Protocatechuic Acid Does Not Affect DHA-Specific Transcriptional Effects on Lipid Metabolism in Cultured Hepatic Cells" Nutrients 12, no. 10: 2952. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12102952