Antibiotic Resistance of Bacteria in Two Marine Mammal Species, Harbor Seals and Harbor Porpoises, Living in an Urban Marine Ecosystem, the Salish Sea, Washington State, USA
Abstract
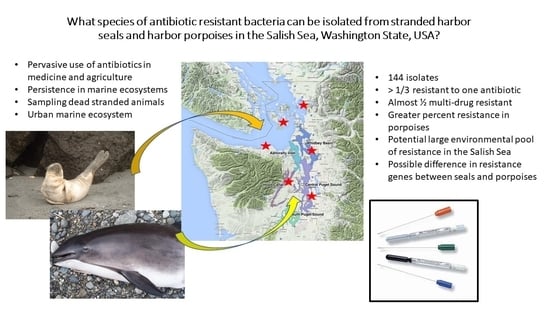
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Study Area
2.3. Sample Collection
2.4. Bacterial Isolation and Antimicrobial Sensitivity Testing
2.5. Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Antibiotic Resistance. Available online: http://www.who.int/news-room/factsheets/detail/antibiotic-resistance (accessed on 27 May 2020).
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. Combating Antimicrobial Resistance: A One Health Approach to a Global Threat: Proceedings of a Workshop; The National Academies Press: Washington, DC, USA, 2017. [Google Scholar]
- Smith, D.L.; Harris, A.D.; Johnson, J.A.; Silbergeld, E.K.; Morris, J.J.G. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 6434–6439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, J.R.; Gast, R.J.; Fujioka, R.S.; Solo-Gabriele, H.M.; Meschke, J.S.; Amaral-Zettler, L.; Del Castillo, E.; Polz, M.F.; Collier, T.K.; Strom, M.; et al. The coastal environment and human health: Microbial indicators, pathogens, sentinels and reservoirs. Environ. Health 2008, 7, S3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial Food Animal Production, Antimicrobial Resistance, and Human Health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Johnson, S.P.; Nolan, S.; Gulland, F.M. Antimicrobial susceptibility of bacteria isolated from pinnipeds stranded in central and northern California. J. Zoo Wildl. Med. 1998, 29, 288–294. [Google Scholar]
- Miranda, C.D.; Zemelman, R. Antibiotic resistance in fish from the Concepción Bay, Chile. Mar. Pollut. Bull. 2001, 42, 1096–1102. [Google Scholar] [CrossRef]
- Greig, T.W.; Bemiss, J.A.; Lyon, B.R.; Bossart, G.D.; Fair, P.A. Prevalence and diversity of antibiotic-resistant Escherichia coli in bottlenose dolphins (Tursiops truncatus) from the Indian River Lagoon, Florida and Charleston Harbor area, South Caro-lina. Aquat. Mamm. 2007, 33, 185–194. [Google Scholar] [CrossRef]
- Schaefer, A.M.; Goldstein, J.D.; Reif, J.S.; Fair, P.A.; Bossart, G.D. Antibiotic-resistant organisms cultured from Atlantic bot-tlenose dolphins (Tursiops truncatus) inhabiting estuarine waters of Charleston, SC, and Indian River Lagoon, FL. Ecohealth 2009, 6, 33–41. [Google Scholar] [CrossRef]
- Blackburn, J.K.; Mitchell, M.A.; Blackburn, M.-C.H.; Curtis, A.; Thompson, B.A. Evidence of Antibiotic Resistance in Free-Swimming, Top-Level Marine Predatory Fishes. J. Zoo Wildl. Med. 2010, 41, 7–16. [Google Scholar] [CrossRef]
- Stewart, J.R.; Townsend, F.; Lane, S.M.; Dyar, E.; Hohn, A.; Rowles, T.K.; Staggs, L.; Wells, R.S.; Balmer, B.C.; Schwacke, L.H. Survey of antibiotic-resistant bacteria isolated from bottlenose dolphins Tursiops truncatus in the southeastern USA. Dis. Aquat. Org. 2014, 108, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Oates, S.C.; Miller, M.A.; Byrne, B.A.; Chouicha, N.; Hardin, D.; Jessup, D.; Dominik, C.; Roug, A.; Schriewer, A.; Jang, S.S.; et al. Epidemiology and potential land-sea transfer of enteric bacteria from terrestrial to marine species in the monterey bay region of california. J. Wildl. Dis. 2012, 48, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban waste water treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatosy, S.M.; Martiny, A.C. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [Green Version]
- Lockwood, S.K.; Chovan, J.L.; Gaydos, J.K. Aerobic bacterial isolations from harbor seals (Phoca vitulina) stranded in Wash-ington: 1992–2003. J. Zoo Wildl. Med. 2006, 37, 281–291. [Google Scholar] [CrossRef]
- Steiger, G.H.; Calambokidis, J.; Cubbage, J.C.; Skilling, D.E.; Smith, A.W.; Gribble, D.H. Mortality of harbor seal pups at different sites in the inland waters of washington. J. Wildl. Dis. 1989, 25, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Raverty, S.A.; Rhodes, L.D.; Zabek, E.; Eshghi, A.; Cameron, C.E.; Hanson, M.B.; Schroeder, J.P. Respiratory microbiome of endangered southern resident killer whales and microbiota of surrounding sea surface microlayer in the eastern North Pa-cific. Sci. Rep. 2017, 7, 394. [Google Scholar] [CrossRef]
- Melendez, D.; Roberts, M.C.; Greninger, A.L.; Weissman, S.; No, D.; Rabinowitz, P.; Wasser, S. Whole-genome analysis of extraintestinal pathogenic Escherichia coli (ExPEC) MDR ST73 and ST127 isolated from endangered southern resident killer whales (Orcinus orca). J. Antimicrob. Chemother. 2019, 74, 2176–2180. [Google Scholar] [CrossRef]
- Geraci, J.R.; Lounsbury, V.J. Marine Mammals Ashore: A Field Guide for Strandings, 2nd ed.; E. John Schmitz & Sons. Inc: Sparks, MD, USA, 2005; pp. 177–178. [Google Scholar]
- Norman, S.A.; Bowlby, C.E.; Brancato, M.S.; Calambokidis, J.; Duffield, D.; Gearin, P.J.; Gornall, T.A.; Gosho, M.E.; Hanson, B.; Hodder, J.; et al. Cetacean strandings in Oregon and Washington between 1930 and 2002. J. Cetacean Res. Manag. 2004, 6, 87–99. [Google Scholar]
- Warlick, A.J.; Duffield, D.A.; Lambourn, D.M.; Jeffries, S.J.; Rice, J.M.; Gaydos, J.K.; Huggins, J.L.; Calambokidis, J.; Lahner, L.L.; Olson, J.; et al. Spatio-Temporal Characterization of Pinniped Strandings and Human Interaction Cases in the Pacific Northwest, 1991–2016. Aquat. Mamm. 2018, 44, 299–318. [Google Scholar] [CrossRef] [Green Version]
- Carretta, J.V.; Forney, K.A.; Oleson, E.M.; Weller, D.W.; Lang, A.R.; Baker, J.D.; Muto, M.M.; Hanson, B.; Orr, A.J.; Huber, H.R.; et al. U.S. Pacific Marine Mammal Stock Assessments: 2018. NOAA Technical Memorandum NMFS-SWFSC-617; U.S. De-partment of Commerce: Washington, DC, USA, 2019.
- Cottrell, P.E.; Jeffries, S.; Beck, B.; Ross, P.S. Growth And Development In Free-Ranging Harbor Seal (Phoca Vitulina) Pups From Southern British Columbia, Canada. Mar. Mammal Sci. 2002, 18, 721–733. [Google Scholar] [CrossRef]
- Gearin, P.J.; Melin, S.R.; Delong, R.L.; Kajimura, H.; Johnson, M.A. Harbor porpoise interactions with a chinook salmon set-net fishery in Washington State. Rep. Int. Whal. Commn. Special 1994, 15, 427–438. [Google Scholar]
- Krogh, J.; Ianson, D.; Hamme, R.C.; Lowe, C.J. Risks of hypoxia and acidification in the high energy coastal environment near Victoria, Canada’s untreated municipal sewage outfalls. Mar. Pollut. Bull. 2018, 133, 517–531. [Google Scholar] [CrossRef]
- Bonham, P.A. Swab cultures for diagnosing wound infections: A literature review and clinical guideline. J. Wound Ostomy. Cont. 2009, 36, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J.; Markey, B.K.; Leonard, F.C.; FitzPatrick, E.S.; Fanning, S.; Hartigan, P.J. Veterinary Microbiology and Microbial Disease, 2nd ed.; Wiley-Blackwell: West Sussex, UK, 2011. [Google Scholar]
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying definitions for multidrug resistance, extensive drug re-sistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamina-tion of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Chitanand, M.P.; Kadam, T.A.; Gyananath, G.; Totewad, N.D.; Balhal, D.K. Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian J. Microbiol. 2010, 50, 216–220. [Google Scholar] [CrossRef] [Green Version]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 2015, 22, 10969–10980. [Google Scholar] [CrossRef]
- Tambekar, D.H.; Hirulkar, N.B.; Waghmare, A.S. MAR indexing to discriminate the source of faecal contamination in drinking water. Nat. Environ. Pollut. Tech. 2005, 4, 525–528. [Google Scholar] [CrossRef] [Green Version]
- Kaneene, B.J.; Miller, R.; Sayah, R.; Johnson, Y.J.; Gilliland, D.; Gardiner, J.C. Considerations when using discrimination function analysis of antimicrobial resistance profiles to identify sources of faecal contamination of surface water in Michi-gan. Appl. Environ. Microbiol. 2007, 73, 2878–2890. [Google Scholar] [CrossRef] [Green Version]
- Kulldorff, M. A spatial scan statistic. Commun. Stat. Theory Methods 1997, 26, 1481–1496. [Google Scholar] [CrossRef]
- Smaldone, G.; Marrone, R.; Cappiello, S.; Martin, G.A.; Oliva, G.; Cortesi, M.L.; Anastasio, A. Occurrence of antibiotic re-sistance in bacteria isolated from seawater organisms caught in Campania Region: Preliminary study. BMC Vet. Res. 2014, 10, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.; Schreier, H.J.; Lanska, L.; Hale, M.S. The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramey, A.M.; Ahlstrom, C.A. Antibiotic Resistant Bacteria in Wildlife: Perspectives on Trends, Acquisition and Dissemination, Data Gaps, and Future Directions. J. Wildl. Dis. 2020, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.M.; Gast, R.J.; Bogomolni, A.; Ellis, J.C.; Lentell, B.J.; Touhey, K.; Moore, M. Occurrence and patterns of antibiotic re-sistance in vertebrates off the Northeastern United States coast. FEMS Microbiol. Ecol. 2009, 67, 421–431. [Google Scholar] [CrossRef]
- Schaefer, A.M.; Bossart, G.D.; Harrington, T.; Fair, P.A.; McCarthy, P.J.; Reif, J.S. Temporal Changes in Antibiotic Resistance Among Bacteria Isolated from Common Bottlenose Dolphins (Tursiops truncatus) in the Indian River Lagoon, Florida, 2003–2015. Aquat. Mamm. 2019, 45, 533–542. [Google Scholar] [CrossRef]
- Higgins, R. Bacteria and fungi of marine mammals: A review. Can. Veter. J. 2000, 41, 105–116. [Google Scholar]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Haenni, M.; Bour, M.; Châtre, P.; Madec, J.-Y.; Plésiat, P.; Jeannot, K. Resistance of Animal Strains of Pseudomonas aeruginosa to Carbapenems. Front. Microbiol. 2017, 8, 1847. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2015, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.; Wayne, A.; Fellman, C.L.; Rosenbaum, M.H. Usage patterns of carbapenem antimicrobials in dogs and cats at a veterinary tertiary care hospital. J. Veter. Intern. Med. 2019, 33, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Swenshon, M.; Lammler, C.; Siebert, U. Identification and Molecular Characterization of Beta-Hemolytic Streptococci Isolated from Harbor Porpoises (Phocoena phocoena) of the North and Baltic Seas. J. Clin. Microbiol. 1998, 36, 1902–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, C.C.; Yund, P.O.; Ford, T.E.; Matassa, K.A.; Bass, A.L. Increase in Antimicrobial Resistance in Bacteria Isolated from Stranded Marine Mammals of the Northwest Atlantic. EcoHealth 2013, 10, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Boon, P.I.; Cattanach, M. Antibiotic resistance of native and faecal bacteria isolated from rivers, reservoirs and sewage treatment facilities in Victoria, south-eastern Australia. Lett. Appl. Microbiol. 1999, 28, 164–168. [Google Scholar] [CrossRef] [Green Version]
- Lima-Bittencourt, C.I.; Cursino, L.; Gonçalves-Dornelas, H.; Pontes, D.S.; Nardi, R.M.D.; Callisto, M.; Chartone-Souza, E.; Nascimento, A.M. Multiple antimicrobial resistance in Enterobacteriaceae isolates from pristine freshwater. Genet. Mol. Res. 2007, 6, 510–521. [Google Scholar]
- Coleman, B.L.; Louie, M.; Salvadori, M.I.; McEwen, S.A.; Neumann, N.; Sibley, K.; Irwin, R.J.; Jamieson, F.B.; Daignault, D.; Majury, A.; et al. Contamination of Canadian private drinking water sources with antimicrobial resistant Escherichia coli. Water Res. 2013, 47, 3026–3036. [Google Scholar] [CrossRef]
- Lyimo, B.; Buza, J.; Subbiah, M.; Smith, W.A.; Call, D.R. Comparison of antibiotic resistant Escherichia coli obtained from drinking water sources in northern Tanzania: A cross-sectional study. BMC Microbiol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pantak, S.P.; Gopal, K. Prevalence of bacterial contamination with antibiotic resistant and enterotoxigenic fecal coliforms in treated drinking water. J. Toxicol. Environ. Health A 2008, 71, 427–433. [Google Scholar] [CrossRef]
- Nguyen, C.C.; Hugie, C.N.; Kile, M.L.; Navab-Daneshmand, T. Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: A review. Front. Environ. Sci. Eng. 2019, 13, 46. [Google Scholar] [CrossRef]
- Akmajian, A.; Calambokidis, J.; Huggins, J.L.; Lambourn, D.M. Age, region, and temporal patterns of trace elements meas-ured in stranded harbor seals (Phoca vitulina richardii) from Washington inland waters. Northwest Nat. 2014, 95, 83–91. [Google Scholar] [CrossRef]
- Ashley, E.A.; Olson, J.K.; Raverty, S.; Wilkinson, K.; Gaydos, J.K. Trace Element Concentrations in Livers of Pacific Harbor Seals (Phoca vitulina richardii) from San Juan County, Washington, USA. J. Wildl. Dis. 2020, 56, 429. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.; Delorme, T.; Nasr, P. Patient age as a factor of antibiotic resistance in methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2017, 66, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Miagkoff, L.; Archambault, M.; Bonilla, A.G. Antimicrobial susceptibility patterns of bacterial isolates cultured from syno-vial fluid samples from horses with suspected septic synovitis: 108 cases (2008–2017). J. Am. Vet. Med. Assoc. 2020, 256, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.A.; Holekamp, K.E.; Winters, A.D.; Theis, K.R. Body site-specific microbiota reflect sex and age-class among wild spotted hyenas. FEMS Microbiol. Ecol. 2020, 96, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Editorial: Antibiotic Resistance in Aquatic Systems. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Frey, E. The role of companion animal veterinarians in one-health efforts to combat antimicrobial resistance. J. Am. Veter. Med. Assoc. 2018, 253, 1396–1404. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treat-ment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Castillo, F.Y.; Harel, J.; Moreno-Flores, A.C.; Loera-Muro, A.; Guerrero-Barrera, A.L.; Avelar-González, F.J. Anti-microbial resistance: The role of aquatic environments. Int. J. Curr. Res. Acad. Rev. 2014, 2, 231–246. [Google Scholar]
- Gaydos, J.K.; Dierauf, L.; Kirby, G.; Brosnan, D.; Gilardi, K.; Davis, G.E. Top 10 principles for designing healthy coastal eco-systems like the Salish Sea. Ecohealth 2008, 5, 460. [Google Scholar] [CrossRef] [Green Version]
- Fresh, K.; Dethier, M.; Simenstad, C.; Logsdon, M.; Shipman, H.; Tanner, C.D.; Leschine, T.M.; Mumford, T.F.; Gelfenbaum, G.; Shuman, R.; et al. Implications of observed anthropogenic changes to the nearshore ecosystems in Puget Sound. Puget Sound Nearshore Ecosystem Restoration Project. Tech. Rep. 2011. Available online: http://www.pugetsoundnearshore.org/technical_papers/implications_of_observed_ns_change.pdf (accessed on 24 June 2020).
- Lamb, J.B.; Van De Water, J.A.J.M.; Bourne, D.G.; Altier, C.; Hein, M.Y.; Fiorenza, E.A.; Abu, N.; Jompa, J.; Harvell, C.D. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 2017, 355, 731–733. [Google Scholar] [CrossRef]
- Shelton, A.O.; Francis, T.B.; Feist, B.E.; Williams, G.D.; Lindquist, A.; Levin, P.S. Forty years of seagrass population stability and resilience in an urbanizing estuary. J. Ecol. 2016, 105, 458–470. [Google Scholar] [CrossRef]
- Bossart, G.D. Marine Mammals as Sentinel Species for Oceans and Human Health. Veter. Pathol. 2010, 48, 676–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Gast, R.J.; Bogomolni, A.L. Marine vertebrate zoonoses: An overview of the DAO Special Issue. Dis. Aquat. Organ. 2008, 81, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waltzek, T.B.; Cortés-Hinojosa, G.; Wellehan, J.F.X., Jr.; Gray, G.C. Marine mammal zoonoses: A review of disease manifes-tations. Zoonoses Public Health 2012, 59, 521–535. [Google Scholar] [CrossRef]
- US Centers for Disease Control and Prevention (CDC), National Notifiable Diseases Surveillance System. Available online: https://wwwn.cdc.gov/nndss/ (accessed on 26 May 2020).
- Paranjpye, R.N.; Nilsson, W.; Liermann, M.; Hilborn, E.D.; George, B.J.; Li, Q.; Bill, B.D.; Trainer, V.L.; Strom, M.S.; Sandifer, P.A. Environmental influences on the seasonal distribution of Vibrio parahaemolyticus in the Pacific Northwest of the USA. FEMS Microbiol. Ecol. 2012, 91, fiv121. [Google Scholar] [CrossRef] [Green Version]
- Strom, M.S.; Paranjpye, R.N. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2000, 2, 177–188. [Google Scholar] [CrossRef]
- Kaysner, C.A.; Abeyta, C.; Wekell, M.M.; DePaola, A.; Stott, R.F.; Leitch, J.M. Incidence of Vibrio cholerae from estuaries of the United States West Coast. Appl. Environ. Microbiol. 1987, 53, 1344–1348. [Google Scholar] [CrossRef] [Green Version]
- Hunt, T.; Ziccardi, M.; Gulland, F.; Yochem, P.; Hird, D.; Rowles, T.; Mazet, J. Health risks for marine mammal workers. Dis. Aquat. Org. 2008, 81, 81–92. [Google Scholar] [CrossRef]
- Tryland, M. Zoonoses of arctic marine mammals. Infect. Dis. Rev. 2000, 2, 55–64. [Google Scholar]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-M.; Holmes, E.C.; Chen, X.; Tian, J.-H.; Lin, X.-D.; Qin, X.-C.; Gao, W.-H.; Liu, J.; Wu, Z.; Zhang, Y.-Z. Diverse and abundant resistome in terrestrial and aquatic vertebrates revealed by transcriptional analysis. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]




| Organism | Number of Isolates | |
|---|---|---|
| Phoca vitulina | Phocoena phocoena | |
| Acinetobacter spp. | 0 | 1 (1/0) |
| Actinomyces | 1 (N/A) | 0 |
| Aeromonas hydrophila | 2 (2/1) | 3 (3/2) |
| Arcanobacterium haemolyticum | 1 (N/A) | 0 |
| Arcanobacterium phocae | 2 (2/0) | 0 |
| β-hemolytic Streptococcus | 8 (8/8) | 1 (1/1) |
| Buttiauxella agrestis | 0 | 2 (0/0) |
| Campylobacter spp. | 2 (N/A) | 0 |
| Escherichia coli | 72 (8/4) | 10 (1/1) |
| Edwardsiella hoshinae | 1 (0/0) | 0 |
| Edwardsiella tarda | 4 (0/0) | 0 |
| Elizabethkingia meningoseptica | 1 (1/1) | 0 |
| Enterobacter spp. | 1 (1/1) | 0 |
| Escherichia fergusonii | 2 (2/2) | 0 |
| Gamma (γ) hemolytic Streptococcus | 1 (0/0) | 0 |
| Gardnerella vaginalis | 1 (1/0) | 0 |
| Granulicatella adiacens | 1 (N/A) | 0 |
| Hafnia alvei | 1 (1/1) | 0 |
| Moellerella wisconsensis | 0 | 1 (N/A) |
| Pantoea agglomerans | 1 (1/1) | 0 |
| Pasteurella group | 1 (0/0) | 0 |
| Pasteurella multocida | 1 (N/A) | 0 |
| Photobacterium damselae | 0 | 1 (0/0) |
| Plesiomonas shigelloides | 1 (1/0) | 1 (1/0) |
| Proteus penneri | 0 | 1 (1/1) |
| Proteus vulgaris | 1 (1/1) | 0 |
| Providencia rettgeri | 0 | 1 (1/1) |
| Pseudomonas aeruginosa | 5 (5/5) | 0 |
| Pseudomonas fluorescens | 0 | 2 (2/2) |
| Salmonella spp. | 3 (3/3) | 0 |
| Serratia fonticola | 1 (0/0) | 0 |
| Shewanella algae | 2 (1/0) | 2 (2/2) |
| Shewanella putrefaciens | 0 | 1 (1/1) |
| Vibrio alginolyticus | 0 | 2 (2/1) |
| Vibrio cholera | 0 | 1 (0/0) |
| Vibrio fluvialis | 0 | 1 (1/0) |
| Vibrio parahaemolyticus | 0 | 2 (2/0) |
| Vibrio vulnificus | 1 (1/1) | 0 |
| Total | 117 | 33 |
| Source | Number of Isolates | |
|---|---|---|
| Phoca vitulina | Phocoena phocoena | |
| Abscess (brain) | 1 (1/1) | 0 |
| Abscess (hind flipper) | 2 (1/1) | 0 |
| Bronchiole | 1 (N/A) | 0 |
| Colostrum | 2 (1/1) | 2 (2/2) |
| Kidney | 1 (0/0) | 2 (1/0) |
| Large intestine | 81 (18/12) | 22 (12/8) |
| Lung parenchyma | 13 (7/7) | 0 |
| Lymph node (unspecified) | 2 (2/2) | 0 |
| Lymph node (mediastinal) | 0 | 2 (2/1) |
| Nares | 1 (0/0) | 0 |
| Oral ulcers | 1 (1/0) | 0 |
| Peritoneal fluid | 9 (3/2) | 0 |
| Pleura | 0 | 2 (1/1) |
| Scapular joint | 1 (1/0) | 0 |
| Thoracic cavity | 0 | 2 (1/1) |
| Tonsil | 0 | 2 (1/0) |
| Uterus | 2 (1/1) | 0 |
| Total | 117 | 34 |
| Bacteria | Gram Stain | Number Isolates | AM | AC | AX | CP | CF | CV | CR | CH | DX | EN | FL | GE | IM | MA | TMS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phoca vitulina | |||||||||||||||||
| Escherichia coli | N | 72 | 4 | 1 | 5 | 1 | 1 | 1 | 1 | 3 | |||||||
| Beta-hemolytic Streptococcus | P | 8 | 8 | 8 | |||||||||||||
| Pseudomonas aeruginosa | N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 1 | 5 | 5 | ||||||
| Salmonella spp. | N | 3 | 3 | 3 | 3 | ||||||||||||
| Aeromonas hydrophila | N | 2 | 2 | 2 | 1 | ||||||||||||
| Shewanella algae | N | 2 | 1 | ||||||||||||||
| Phocoena phocoena | |||||||||||||||||
| Aeromonas hydrophila | N | 3 | 3 | 2 | 1 | ||||||||||||
| Pseudomonas fluorescens | N | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | |||||
| Shewanella algae | N | 2 | 1 | 2 | 2 | 1 | |||||||||||
| Vibrio alginolyticus | N | 2 | 1 | 1 | 1 | ||||||||||||
| Vibrio parahaemolyticus | N | 2 | 2 | ||||||||||||||
| Total | 103 | 3 | 3 | 20 | 17 | 7 | 7 | 9 | 8 | 9 | 1 | 16 | 4 | 2 | 9 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norman, S.A.; Lambourn, D.M.; Huggins, J.L.; Gaydos, J.K.; Dubpernell, S.; Berta, S.; Olson, J.K.; Souze, V.; Evans, A.; Carlson, B.; et al. Antibiotic Resistance of Bacteria in Two Marine Mammal Species, Harbor Seals and Harbor Porpoises, Living in an Urban Marine Ecosystem, the Salish Sea, Washington State, USA. Oceans 2021, 2, 86-104. https://0-doi-org.brum.beds.ac.uk/10.3390/oceans2010006
Norman SA, Lambourn DM, Huggins JL, Gaydos JK, Dubpernell S, Berta S, Olson JK, Souze V, Evans A, Carlson B, et al. Antibiotic Resistance of Bacteria in Two Marine Mammal Species, Harbor Seals and Harbor Porpoises, Living in an Urban Marine Ecosystem, the Salish Sea, Washington State, USA. Oceans. 2021; 2(1):86-104. https://0-doi-org.brum.beds.ac.uk/10.3390/oceans2010006
Chicago/Turabian StyleNorman, Stephanie A., Dyanna M. Lambourn, Jessica L. Huggins, Joseph K. Gaydos, Sandra Dubpernell, Susan Berta, Jennifer K. Olson, Victoria Souze, Alysha Evans, Betsy Carlson, and et al. 2021. "Antibiotic Resistance of Bacteria in Two Marine Mammal Species, Harbor Seals and Harbor Porpoises, Living in an Urban Marine Ecosystem, the Salish Sea, Washington State, USA" Oceans 2, no. 1: 86-104. https://0-doi-org.brum.beds.ac.uk/10.3390/oceans2010006