Epidemiology, Biotic Interactions and Biological Control of Armillarioids in the Northern Hemisphere
Abstract
:1. Introduction
2. Identification
3. Biodiversity, Population Genetics
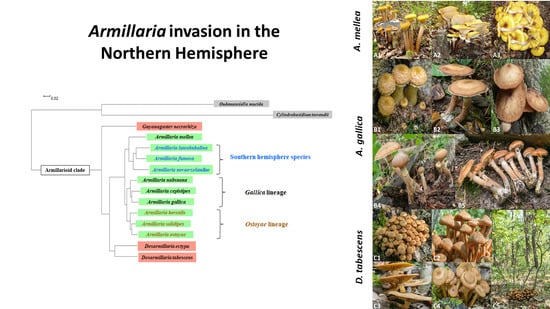
4. Distribution and Host Range of Pathogenic Armillarioid Species and Lineages
5. Biology and Infection Strategies of Armillarioid Species
5.1. Transmission and Infection Pathways
5.2. Penetration, Colonization and Disease Development
5.3. Susceptibility of the Host and Plant Defense Mechanisms Associated with Armillaria Infection
6. Biotic Factors Facilitating Armillaria Transmission and Infection
6.1. Fungi Stimulating Armillaria Infection
6.2. Interactions between Armillarioids and Insects
6.3. The Role of Forest Condition in Armillarioid Infection
7. Towards the Biological Control of Armillarioid Root Disease
7.1. Bacteria
7.2. Fungi
7.3. Nematodes
7.4. Substances Derived from Cyanobacteria and Plants
7.5. Combined Approaches
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heinzelmann, R.; Dutech, C.; Tsykun, T.; Labbé, F.; Soularue, J.-P.; Prospero, S. Latest advances and future perspectives in Armillaria research. Can. J. Plant Pathol. 2019, 41, 1–23. [Google Scholar] [CrossRef]
- Dettman, J.R.; van der Kamp, B.J. The population structure of Armillaria ostoyae in the southern interior of British Columbia. Can. J. Bot. 2001, 79, 612–620. [Google Scholar] [CrossRef]
- Rishbeth, J. Species of Armillaria in southern England. Plant Pathol. 1982, 31, 9–17. [Google Scholar] [CrossRef]
- Mwenje, E.; Wingfield, B.D.; Coetzee, M.P.; Wingfield, M.J. Molecular characterisation of Armillaria species from Zimbabwe. Mycol. Res. 2003, 107, 291–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Żółciak, A. Armillaria species in coniferous stands. Acta Mycol. 2007, 42, 211–217. [Google Scholar] [CrossRef]
- Tigang, S.P.; Tchoumi, J.M.T.; Roux, J.; Nguefack, J.; Boyogueno, A.D.B.; Mbenoun, M.; Mfegue, C.V.; Nyassé, S.; Nkeng, M.N.; ten Hoopen, G.M. Armillaria root rot threatens Cameroon’s Penja pepper (Piper nigrum L.). Trop. Plant Pathol. 2020, 45, 534–543. [Google Scholar] [CrossRef]
- Worrall, J.J.; Egeland, L.; Eager, T.; Mask, R.A.; Johnson, E.W.; Kemp, P.A.; Shepperd, W.D. Rapid mortality of Populus tremuloides in southwestern Colorado, USA. For. Ecol. Manag. 2008, 255, 686–696. [Google Scholar] [CrossRef] [Green Version]
- Wingfield, M.J.; Slippers, B.; Wingfield, B.D. 2010. Novel associations between pathogens, insects and tree species threaten world forests. N. Z. J. For. Sci. 2010, 40, 95–103. [Google Scholar]
- Lushaj, B.M.; Woodward, S.; Keča, N.; Intini, M. Distribution, ecology and host range of Armillaria species in Albania. For. Pathol. 2010, 40, 485–499. [Google Scholar] [CrossRef]
- Cruickshank, M.G. Yield reduction in spruce infected with Armillaria solidipes in the southern interior of British Columbia. For. Pathol. 2011, 41, 425–428. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Slaughter, G.W. Root disease and canopy gaps in developed areas of Yosemite Valley, California. For. Ecol. Manag. 2001, 146, 159–167. [Google Scholar] [CrossRef]
- Cruickshank, M.G.; Morrison, D.J.; Lalumière, A. The interaction between competition in interior Douglas-fir plantations and disease caused by Armillaria ostoyae in British Columbia. For. Ecol. Manag. 2009, 257, 443–452. [Google Scholar] [CrossRef]
- Chandelier, A.; Gerarts, F.; San Martin, G.; Herman, M.; Delahaye, L. Temporal evolution of collar lesions associated with ash dieback and the occurrence of Armillaria in Belgian forests. For. Pathol. 2016, 46, 289–297. [Google Scholar] [CrossRef]
- Pavlov, I.N. Biotic and abiotic factors as causes of coniferous forests dieback in Siberia and Far East. Contemp. Probl. Ecol. 2015, 8, 440–456. [Google Scholar] [CrossRef]
- Cruickshank, M.G.; Filipescu, C.N. Allometries of coarse tree, stem, and crown measures in Douglas-fir are altered by Armillaria root disease. Botany 2012, 90, 711–721. [Google Scholar] [CrossRef]
- Thomas, F.M.; Blank, R.; Hartmann, G. Abiotic and biotic factors and their interactions as causes of oak decline in Central Europe. For. Pathol. 2002, 32, 277–307. [Google Scholar] [CrossRef]
- Baumgartner, K.; Rizzo, D.M. Spread of Armillaria root disease in a California vineyard. Am. J. Enol. Viticult. 2002, 53, 197–203. [Google Scholar]
- Baumgartner, K. Root collar excavation for postinfection control of Armillaria root disease of grapevine. Plant Dis. 2004, 88, 1235–1240. [Google Scholar] [CrossRef] [Green Version]
- Raabe, R. Host list of the root rot fungus, Armillaria mellea. Hilgardia 1962, 33, 25–88. [Google Scholar] [CrossRef]
- Sicoli, G.; Annese, V.; De Gioia, T.; Luisi, N. Armillaria pathogenicity tests on oaks in southern Italy. J. Plant Pathol. 2002, 84, 107–111. [Google Scholar] [CrossRef]
- Kromroy, K.W.; Blanchette, R.A.; Grigal, D.F. Armillaria species on small woody plants, small woody debris, and root fragments in red pine stands. Can. J. For. Res. 2005, 35, 1487–1495. [Google Scholar] [CrossRef] [Green Version]
- Baumgartner, K.; Coetzee, M.P.; Hoffmeister, D. Secrets of the subterranean pathosystem of Armillaria. Mol. Plant Pathol. 2011, 12, 515–534. [Google Scholar] [CrossRef] [PubMed]
- Guillaumin, J.J.; Legrand, P. Armillaria root rots. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CAB International: Boston, MA, USA, 2013; pp. 159–177. [Google Scholar] [CrossRef]
- Chapman, B.; Schellenberg, B. An exploration of ringbarking to reduce the severity of Armillaria root disease in logged areas in British Columbia. Can. J. For. Res. 2015, 45, 1803–1805. [Google Scholar] [CrossRef] [Green Version]
- Kile, G.A.; McDonald, G.I.; Byler, J.W. Ecology and disease in natural forests. In Armillaria Root Disease; Agriculture Handbooks AH691; Shaw, C.G., III, Kile, G.A., Eds.; USDA: Washington, DC, USA, 1991; pp. 102–121. [Google Scholar]
- Mihail, J.D.; Obert, M.; Bruhn, J.N.; Taylor, S.J. Fractal geometry of diffuse mycelia and rhizomorphs of Armillaria species. Mycol. Res. 1995, 99, 81–88. [Google Scholar] [CrossRef]
- Korhonen, K. Interfertility and clonal size in the Armillariella mellea complex. Karstenia 1978, 18, 31–42. [Google Scholar] [CrossRef]
- Kile, G.A. Identification of genotypes and the clonal development of Armillaria luteobubalina Watling & Kile in eucalypt forests. Aust. J. Bot. 1983, 31, 657–671. [Google Scholar] [CrossRef]
- Ullrich, R.C.; Anderson, J.B. Sex and diploidy in Armillaria mellea. Exp. Mycol. 1978, 2, 119–129. [Google Scholar] [CrossRef]
- Anderson, J.B.; Ullrich, R.C.; Roth, L.F.; Filip, G.M. Genetic identification of clones of Armillaria mellea in coniferous forests in Washington. Phytopathology 1979, 69, 1109–1111. [Google Scholar] [CrossRef]
- Thompson, W. Distribution, development and functioning of mycelia cord systems of decomposer basidiomycetes of the deciduous woodland floor. In The Ecology and Physiology of the Fungal Mycelium; Jennings, D.H., Rayner, A.D.M., Eds.; Cambridge University Press: Cambridge, UK, 1984; pp. 185–214. [Google Scholar]
- Rizzo, D.M.; Harrington, T.C. Delineation and biology of clones of Armillaria ostoyae, A. gemina and A. calvescens. Mycologia 1993, 85, 164–174. [Google Scholar] [CrossRef]
- Bragaloni, M.; Anselmi, N.; Cellerino, G.P. Identification of European Armillaria species by analysis of isozyme profiles. Eur. J. For. Pathol. 1997, 27, 147–157. [Google Scholar] [CrossRef]
- Bruhn, J.N. Identification of Armillaria field isolates using isozymes and mycelial growth characteristics. Mycopathologia 1998, 142, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Burdsall, H.H., Jr.; Banik, M.; Cook, M.E. Serological differentiation of three species of Armillaria and Lentinula edodes by enzyme-linked immunosorbent assay using immunized chickens as a source of antibodies. Mycologia 1990, 82, 415–423. [Google Scholar] [CrossRef]
- Keča, N.; Bodles, W.J.; Woodward, S.; Karadžić, D.; Bojović, S. Molecular-based identification and phylogeny of Armillaria species from Serbia and Montenegro. For. Pathol. 2006, 36, 41–57. [Google Scholar] [CrossRef]
- Schulze, S.; Bahnweg, G.; Möller, E.M.; Sandermann, H. Identification of the genus Armillaria by specific amplification of an rDNA-ITS fragment and evaluation of genetic variation within A. ostoyae by rDNA-RFLP and RAPD analysis. Eur. J. For. Pathol. 1997, 27, 225–239. [Google Scholar] [CrossRef]
- Smith-White, J.L.; Summerell, B.A.; Gunn, L.V.; Rinzin, C.; Porter, C.; Burgess, L.W. Molecular detection and differentiation of Australian Armillaria species. Australas. Plant Path. 2002, 31, 75–79. [Google Scholar] [CrossRef]
- Fox, R.T.V. Managing Armillaria root rot. J. Food Agric. Environ. 2003, 1, 95–100. [Google Scholar]
- Schena, L.; Nigro, F.; Ippolito, A. Real-time PCR detection and quantification of soilborne fungal pathogens: The case of Rosellinia necatrix, Phytophthora nicotianae, P. citrophthora, and Verticillium dahliae. Phytopathol. Mediterr. 2004, 43, 273–280. [Google Scholar] [CrossRef]
- Longa, C.M.; La Porta, N. Rapid identification of Armillaria species by PCR–DGGE. J. Microbiol. Meth. 2014, 107, 63–65. [Google Scholar] [CrossRef]
- Frontz, T.M.; Davis, D.D.; Bunyard, B.A.; Royse, D.J. Identification of Armillaria species isolated from bigtooth aspen based on rDNA RFLP analysis. Can. J. For. Res. 1998, 28, 141–149. [Google Scholar] [CrossRef]
- Johannesson, H.; Stenlid, J. Molecular identification of wood-inhabiting fungi in an unmanaged Picea abies forest in Sweden. For. Ecol. Manag. 1999, 115, 203–211. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.N.; Henricot, B. Identification of fungal species beyond morphology. Mycologist 2002, 16, 42–46. [Google Scholar] [CrossRef]
- Coetzee, M.P.; Wingfield, B.D.; Bloomer, P.; Ridley, G.S.; Wingfield, M.J. Molecular identification and phylogeny of Armillaria isolates from South America and Indo-Malaysia. Mycologia 2003, 95, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, M.A.; Wingfield, B.D.; Roux, J.; Crous, P.W.; Denman, S.; Wingfield, M.J. Discovery of two Northern Hemisphere Armillaria species on Proteaceae in South Africa. Plant Pathol. 2003, 52, 604–612. [Google Scholar] [CrossRef] [Green Version]
- Sicoli, G.; Fatehi, J.; Stenlid, J. Development of species-specific PCR primers on rDNA for the identification of European Armillaria species. For. Pathol. 2003, 33, 287–297. [Google Scholar] [CrossRef]
- Fukuda, M.; Nakashima, E.; Hayashi, K.; Nagasawa, E. Identification of the biological species of Armillaria associated with Wynnea and Entoloma abortivum using PCR-RFLP analysis of the intergenic region (IGR) of ribosomal DNA. Mycol. Res. 2003, 107, 1435–1441. [Google Scholar] [CrossRef]
- Coetzee, M.P.; Wingfield, B.D.; Kirisits, T.; Chhetri, D.B.; Bloomer, P.; Wingfield, M.J. Identification of Armillaria isolates from Bhutan based on DNA sequence comparisons. Plant Pathol. 2005, 54, 36–45. [Google Scholar] [CrossRef]
- Escofet Crespo, P.E.; Aguín Casal, O.; Mansilla Vázquez, J.P. Detection and identification with molecular techniques of species of the genus Armillaria from soil samples. Bol. San. Veg. Plagas 2006, 32, 231–240. [Google Scholar]
- Anderson, J.B.; Bailey, S.S.; Pukkila, P.J. Variation in ribosomal DNA among biological species of Armillaria, a genus of root-infecting fungi. Evolution 1989, 43, 1652–1662. [Google Scholar] [CrossRef]
- Anderson, J.B.; Petsche, D.M.; Smith, M.L. Restriction fragment polymorphisms in biological species of Armillaria mellea. Mycologia 1987, 79, 69–76. [Google Scholar] [CrossRef]
- Kim, M.S.; Klopfenstein, N.B.; McDonald, G.I.; Arumuganathan, K.; Vidaver, A.K. Characterization of North American Armillaria species by nuclear DNA content and RFLP analysis. Mycologia 2000, 92, 874–883. [Google Scholar] [CrossRef] [Green Version]
- Harrington, T.C.; Wingfield, B.D. A PCR-based identification method for species of Armillaria. Mycologia 1995, 87, 280–288. [Google Scholar] [CrossRef]
- Gezahgne, A.; Coetzee, M.P.; Wingfield, B.D.; Wingfield, M.J.; Roux, J. Identification of the Armillaria root rot pathogen in Ethiopian plantations. For. Pathol. 2004, 34, 133–145. [Google Scholar] [CrossRef]
- Coetzee, M.P.; Wingfield, B.D.; Harrington, T.C.; Steimel, J.; Coutinho, T.A.; Wingfield, M.J. The root rot fungus Armillaria mellea introduced into South Africa by early Dutch settlers. Mol. Ecol. 2001, 10, 387–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Klopfenstein, N.B.; Hanna, J.W.; McDonald, G.L. Characterization of North American Armillaria species: Genetic relationships determined by ribosomal DNA sequences and AFLP markers. For. Pathol. 2006, 36, 145–164. [Google Scholar] [CrossRef]
- Hanna, J.W.; Klopfenstein, N.B.; Kim, M.S.; McDonald, G.I.; Moore, J.A. Phylogeographic patterns of Armillaria ostoyae in the western United States. For. Pathol. 2007, 37, 192–216. [Google Scholar] [CrossRef]
- Ross-Davis, A.L.; Stewart, J.E.; Hanna, J.W.; Kim, M.S.; Knaus, B.J.; Cronn, R.; Rai, H.; Richardson, B.A.; McDonald, G.I.; Klopfenstein, N.B. Transcriptome of an Armillaria root disease pathogen reveals candidate genes involved in host substrate utilization at the host–pathogen interface. For. Pathol. 2013, 43, 468–477. [Google Scholar] [CrossRef]
- Maphosa, L.; Wingfield, B.D.; Coetzee, M.P.; Mwenje, E.; Wingfield, M.J. Phylogenetic relationships among Armillaria species inferred from partial elongation factor 1-alpha DNA sequence data. Australas. Plant Pathol. 2006, 35, 513–520. [Google Scholar] [CrossRef]
- Hasegawa, E.; Ota, Y.; Hattori, T.; Kikuchi, T. Sequence-based identification of Japanese Armillaria species using the elongation factor-1 alpha gene. Mycologia 2010, 102, 898–910. [Google Scholar] [CrossRef]
- Antonín, V.; Tomšovský, M.; Sedlák, P.; Májek, T.; Jankovský, L. Morphological and molecular characterization of the Armillaria cepistipes–A. gallica complex in the Czech Republic and Slovakia. Mycol. Progr. 2009, 8, 259–271. [Google Scholar] [CrossRef]
- Mulholland, V.; MacAskill, G.A.; Laue, B.E.; Steele, H.; Kenyon, D.; Green, S. Development and verification of a diagnostic assay based on EF-1 α for the identification of Armillaria species in northern Europe. For. Pathol. 2012, 42, 229–238. [Google Scholar] [CrossRef]
- Slobin, L.I. The role of eucaryotic elongation factor Tu in protein synthesis: The measurement of the elongation factor Tu content of rabbit reticulocytes and other mammalian cells by a sensitive radioimmunoassay. Eur. J. Biochem. 1980, 110, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Brazee, N.J.; Hulvey, J.P.; Wick, R.L. Evaluation of partial tef1, rpb2, and nLSU sequences for identification of isolates representing Armillaria calvescens and Armillaria gallica from northeastern North America. Fungal Biol. 2011, 115, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Stewart, J.E.; Ota, Y.; Hanna, J.W.; Ross-Davis, A.L.; Klopfenstein, N.B. Phylogenetic relationships among Northern Hemisphere Armillaria species based on the tef-1 alpha locus. Phytopathology 2012, 102, 63–64. [Google Scholar]
- Tsykun, T.; Rigling, D.; Prospero, S. A new multilocus approach for a reliable DNA-based identification of Armillaria species. Mycologia 2013, 105, 1059–1576. [Google Scholar] [CrossRef]
- Klopfenstein, N.B.; Hanna, J.W.; Ross-Davis, A.L.; Stewart, J.E.; Ota, Y.; Medel-Ortiz, R.; López-Ramírez, M.A.; Elías-Román, R.D.; Alvarado-Rosales, D.; Kim, M.S. Armillaria phylogeny based on tef-1α sequences suggests ongoing divergent speciation within the boreal floristic kingdom. In Proceedings of the 60th Annual Western International Forest Disease Work Conference, Tahoe City, CA, USA, 8–12 October 2012; Browning, J., Palacios, P., Eds.; US Department of Agriculture, Forest Service, Forest Health Technology and Enterprise Team: Fort Collins, CO, USA, 2012; pp. 141–144. [Google Scholar]
- Keča, N.; Klopfenstein, N.B.; Kim, M.S.; Solheim, H.; Woodward, S. Initial characterization of unidentified Armillaria isolate from Serbia using LSU-IGS1 and TEF-1a genes. For. Pathol. 2015, 45, 120–124. [Google Scholar] [CrossRef]
- Klopfenstein, N.B.; Stewart, J.E.; Ota, Y.; Hanna, J.W.; Richardson, B.A.; Ross-Davis, A.L.; Elías-Román, R.D.; Korhonen, K.; Keča, N.; Iturritxa, E.; et al. Insights into the phylogeny of Northern Hemisphere Armillaria: Neighbor-net and Bayesian analyses of translation elongation factor 1-α gene sequences. Mycologia 2017, 109, 75–91. [Google Scholar] [CrossRef]
- Coetzee, M.P.A.; Wingfield, B.D.; Wingfield, M.J. Armillaria root-rot pathogens: Species boundaries and global distribution. Pathogens 2018, 7, 83. [Google Scholar] [CrossRef] [Green Version]
- Jayawardena, R.S.; Hyde, K.D.; Chen, Y.J.; Papp, V.; Palla, B.; Papp, D.; Bhunjun, C.S.; Hurdeal, V.G.; Senwanna, C.; Manawashinge, I.S.; et al. One stop shop IV: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 76-100. Fungal Divers. 2020, 103, 87–218. [Google Scholar] [CrossRef]
- Sipos, G.; Prasanna, A.N.; Walter, M.C.; O’Connor, E.; Bálint, B.; Krizsán, K.; Kiss, B.; Hess, J.; Varga, T.; Slot, J.; et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 2017, 1, 1931–1941. [Google Scholar] [CrossRef] [Green Version]
- Koch, R.A.; Wilson, A.W.; Séné, O.; Henkel, T.W.; Aime, M.C. Resolved phylogeny and biogeography of the root pathogen Armillaria and its gasteroid relative, Guyanagaster. BMC Evol. Biol. 2017, 17, 33. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.B.; Kohn, L.M. Clonality in soilborne, plant pathogenic fungi. Annu. Rev. Phytopathol. 1995, 33, 369–391. [Google Scholar] [CrossRef]
- Smith, M.L.; Bruhn, J.N.; Anderson, J.B. Relatedness and spatial distribution of Armillaria genets infecting red pine seedlings. Phytopathology 1994, 84, 822–829. [Google Scholar] [CrossRef]
- Prospero, S.; Rigling, D.; Holdenrieder, O. Population structure of Armillaria species in managed Norway spruce stands in the Alps. New Phytol. 2003, 158, 365–373. [Google Scholar] [CrossRef]
- Warwell, M.V.; McDonald, G.I.; Hanna, J.W.; Kim, M.-S.; Lalande, B.M.; Stewart, J.E.; Hudak, A.T.; Klopfenstein, N.B. Armillaria altimontana is associated with healthy western white pine (Pinus monticola): Potential in situ biological control of the Armillaria root disease pathogen, A. solidipes. Forests 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Tsykun, T.; Rellstab, C.; Dutech, C.; Sipos, G.; Prospero, S. Comparative assessment of SSR and SNP markers for inferring the population genetic structure of the common fungus Armillaria cepistipes. Heredity 2017, 119, 371–380. [Google Scholar] [CrossRef]
- Travadon, R.; Smith, M.E.; Fujiyoshi, P.; Douhan, G.W.; Rizzo, D.M.; Baumgartner, K. Inferring dispersal patterns of the generalist root fungus Armillaria mellea. New Phytol. 2012, 193, 959–969. [Google Scholar] [CrossRef]
- Wargo, P.M.; Shaw, C.G., III. Armillaria root rot: The puzzle is being solved. Plant Dis. 1985, 69, 826–832. [Google Scholar] [CrossRef]
- Luisi, N.; Sicoli, G.; Lerario, P. Observations on Armillaria occurrence in declining oak woods of southern Italy. Ann. For. Sci. 1996, 53, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Chillali, M.; Idder-Ighili, H.; Guillaumin, J.J.; Mohammed, C.; Escarmant, B.L.; Botton, B. Variation in the ITS and IGS regions of ribosomal DNA among the biological species of European Armillaria. Mycol. Res. 1998, 102, 533–540. [Google Scholar] [CrossRef]
- Heinzelmann, R.; Rigling, D.; Prospero, S. Population genetics of the wood-rotting basidiomycete Armillaria cepistipes in a fragmented forest landscape. Fungal Biol. 2012, 116, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, A.; Prodorutti, D.; Pertot, I. Use of bark mulch pre-inoculated with Trichoderma atroviride to control Armillaria root rot. Crop Prot. 2014, 64, 104–109. [Google Scholar] [CrossRef]
- Labbé, F.; Marcais, B.; Dupouey, J.L.; Bélouard, T.; Capdevielle, X.; Piou, D.; Robin, C.; Dutech, C. Pre-existing forests as sources of pathogens? The emergence of Armillaria ostoyae in a recently planted pine forest. For. Ecol. Manag. 2015, 357, 248–258. [Google Scholar] [CrossRef]
- Heinzelmann, R.; Rigling, D. Mycelial fan formation of three sympatric Armillaria species on excised stem segments of Picea abies. For. Pathol. 2016, 46, 187–199. [Google Scholar] [CrossRef]
- Abomo-Ndongo, S.; Guillaumin, J.J. Somatic incompatibility among African Armillaria isolates. Eur. J. For. Pathol. 1997, 27, 201–206. [Google Scholar] [CrossRef]
- Coetzee, M.P.; Wingfield, B.D.; Bloomer, P.; Wingfield, M.J. Phylogenetic analyses of DNA sequences reveal species partitions amongst isolates of Armillaria from Africa. Mycol. Res. 2005, 109, 1223–1234. [Google Scholar] [CrossRef] [Green Version]
- Otieno, W.; Sierra, A.P.; Termorshuizen, A. Characterization of Armillaria isolates from tea (Camellia sinensis) in Kenya. Mycologia 2003, 95, 160–175. [Google Scholar] [CrossRef]
- Cha, J.Y.; Igarashi, T. Armillaria species associated with Gastrodia elata in Japan. Eur. J. For. Pathol. 1995, 25, 319–326. [Google Scholar] [CrossRef]
- Mohammed, N.C.; Guillaumin, J.J.; Berthelay, S. Armillaria species identified in China and Japan. Mycol. Res. 1994, 98, 607–613. [Google Scholar] [CrossRef]
- Terashima, K.; Cha, J.Y.; Yajima, T.; Igarashi, T.; Miura, K. Phylogenetic analysis of Japanese Armillaria based on the intergenic spacer (IGS) sequences of their ribosomal DNA. Eur. J. For. Pathol. 1998, 28, 11–19. [Google Scholar] [CrossRef]
- Qin, G.F.; Zhao, J.; Korhonen, K. A study on intersterility groups of Armillaria in China. Mycologia 2007, 99, 430–441. [Google Scholar] [CrossRef]
- Elías-Román, R.D.; Guzmán-Plazola, R.A.; Klopfenstein, N.B.; Alvarado-Rosales, D.; Calderón-Zavala, G.; Mora-Aguilera, J.A.; Kim, M.-S.; García-Espinosa, R. Incidence and phylogenetic analyses of Armillaria spp. associated with root disease in peach orchards in the State of Mexico, Mexico. For. Pathol. 2013, 43, 390–401. [Google Scholar] [CrossRef]
- Thormann, M.N.; Myrholm, C.L.; Mallett, K.I. Armillaria sinapina in herbaceous plant material from a peatland in Alberta, Canada. Can. J. Bot. 2001, 79, 643–647. [Google Scholar] [CrossRef]
- Brazee, N.J.; Wick, R.L. Armillaria species distribution and site relationships in Pinus-and Tsuga-dominated forests in Massachusetts. Can. J. For. Res. 2011, 41, 1477–1490. [Google Scholar] [CrossRef]
- Schnabel, G.; Agudelo, P.; Henderson, G.W.; Rollins, P.A. Aboveground root collar excavation of peach trees for Armillaria root rot management. Plant Dis. 2012, 96, 681–686. [Google Scholar] [CrossRef]
- Cleary, M.R.; van der Kamp, B.J.; Morrison, D.J. Pathogenicity and virulence of Armillaria sinapina and host response to infection in Douglas-fir, western hemlock and western redcedar in the southern interior of British Columbia. For. Pathol. 2012, 42, 481–491. [Google Scholar] [CrossRef]
- Shaw, C.G., III; Roth, L.F. Persistence and distribution of a clone of Armillaria mellea in a ponderosa pine forest. Phytopathology 1976, 66, 1210–1213. [Google Scholar] [CrossRef]
- Brazee, N.J.; Wick, R.L. Armillaria species distribution on symptomatic hosts in northern hardwood and mixed oak forests in western Massachusetts. For. Ecol. Manag. 2009, 258, 1605–1612. [Google Scholar] [CrossRef]
- Alveshere, B.C.; McMurtrey, S.; Bennett, P.; Kim, M.-S.; Hanna, J.W.; Klopfenstein, N.B.; Blodgett, J.T.; LeBoldus, J.M. Phylogeography and host range of Armillaria gallica in riparian forests of the northern Great Plains, USA. For. Pathol. 2020, in press. [Google Scholar] [CrossRef]
- Keča, N.; Karadžić, D.; Woodward, S. Ecology of Armillaria species in managed forests and plantations in Serbia. For. Pathol. 2009, 39, 217–231. [Google Scholar] [CrossRef]
- Prospero, S.; Holdenrieder, O.; Rigling, D. Comparison of the virulence of Armillaria cepistipes and Armillaria ostoyae on four Norway spruce provenances. For. Pathol. 2004, 34, 1–14. [Google Scholar] [CrossRef]
- Tsykun, T.; Rigling, D.; Nikolaychuk, V.; Prospero, S. Diversity and ecology of Armillaria species in virgin forests in the Ukrainian Carpathians. Mycol. Progr. 2012, 11, 403–414. [Google Scholar] [CrossRef]
- Drakulic, J.; Gorton, C.; Perez-Sierra, A.; Clover, G.; Beal, L. Associations between Armillaria species and host plants in U.K. gardens. Plant Dis. 2017, 101, 1903–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cromey, M.G.; Drakulic, J.; Beal, E.J.; Waghorn, I.A.G.; Perry, J.N.; Clover, G.R.G. Susceptibility of garden trees and shrubs to Armillaria root rot. Plant Dis. 2020, 104, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Tsopelas, P. Distribution and ecology of Armillaria species in Greece. Eur. J. For. Pathol. 1999, 29, 103–116. [Google Scholar] [CrossRef]
- Grillo, R.; Tirró, A.; Pennisi, A.M.; Agosteo, G.E. Armillaria species in Calabria. Micol. Ital. 1996, 25, 92–100. [Google Scholar]
- Hanna, J.W. Armillaria ostoyae: Genetic Characterization and Distribution in the Western United States. Master’s Thesis, University of Idaho, Moscow, ID, USA, 2005. [Google Scholar]
- Morrison, D.J.; Merler, H.; Norris, D. Species of Armillaria in British Columbia. Can. J. Plant Pathol. 1985, 7, 242–246. [Google Scholar] [CrossRef]
- Klopfenstein, N.B.; Lundquist, J.E.; Hanna, J.W.; Kim, M.S.; McDonald, G.I. First report of Armillaria sinapina, a cause of Armillaria root disease, associated with a variety of forest tree hosts on sites with diverse climates in Alaska. Plant Dis. 2009, 93, 111. [Google Scholar] [CrossRef]
- Bérubé, J.A.; Dessureault, M. Morphological studies of the Armillaria mellea complex: Two new species, A. gemina and A. calvescens. Mycologia 1989, 81, 216–225. [Google Scholar] [CrossRef]
- Anderson, J.B. Biological species of Armillaria in North America: Redesignation of groups IV and VIII and enumeration of voucher strains for other groups. Mycologia 1986, 78, 837–839. [Google Scholar] [CrossRef]
- Roll-Hansen, F. The Armillaria species in Europe: A literature review. Eur. J. For. Pathol. 1985, 15, 22–31. [Google Scholar] [CrossRef]
- Akulova, V.S.; Sharov, V.V.; Aksyonova, A.I.; Putintseva, Y.A.; Oreshkova, N.V.; Feranchuk, S.I.; Kuzmin, D.A.; Pavlov, I.N.; Litovka, Y.A.; Krutovsky, K.V. De novo sequencing, assembly and functional annotation of Armillaria borealis genome. BMC Genom. 2020, 21, 534. [Google Scholar] [CrossRef]
- Munda, A. Research on honey fungus (Armillaria (Fr.: Fr.) Staude) in Slovenia. In Znanje za Gozd. Zbornik ob 50. Obletnici Obstoja in Delovanja Gozdarskega Instituta Slovenije; Slovenian Forestry Institute: Ljubljana, Slovenia, 1997; Volume 1, pp. 211–220. [Google Scholar]
- Hood, I.A.; Redfern, D.B.; Kile, G.A. Armillaria in planted hosts. In Armillaria Root Disease; Agriculture Handbook No. 691; Shaw, C.G., III, Kile, G.A., Eds.; United States Department of Agriculture, Forest Service: Washington DC, USA, 1991; pp. 122–149. [Google Scholar]
- Aguín-Casal, O.; Sáinz-Osés, M.J.; Mansilla-Vázquez, J.P. Armillaria species infesting vineyards in northwestern Spain. Eur. J. Plant Pathol. 2004, 110, 683–687. [Google Scholar] [CrossRef]
- Cox, K.D.; Scherm, H. Interaction dynamics between saprobic lignicolous fungi and Armillaria in controlled environments: Exploring the potential for competitive exclusion of Armillaria on peach. Biol. Control 2006, 37, 291–300. [Google Scholar] [CrossRef]
- Perazzolli, M.; Bampi, F.; Faccin, S.; Moser, M.; De Luca, F.; Ciccotti, A.M.; Velasco, R.; Gessler, C.; Pertot, I.; Moser, C. Armillaria mellea induces a set of defense genes in grapevine roots and one of them codifies a protein with antifungal activity. Mol. Plant Microbe Interact. 2010, 23, 485–496. [Google Scholar] [CrossRef]
- Ford, K.L.; Baumgartner, K.; Henricot, B.; Bailey, A.M.; Foster, G.D. A native promoter and inclusion of an intron is necessary for efficient expression of GFP or mRFP in Armillaria mellea. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, K. Fungi belonging to the genera Heterobasidion and Armillaria in Eurasia. In Fungal Communities in Forest Ecosystems. Materials of Coordination Investigations; Storozhenko, V.G., Krutov, V.I., Eds.; Institut Lesovedeniya RAN, Institut lesa Karel’skogo NTs RAN: Moscow/Petrozavodsk, Russia, 2004; Volume 2, pp. 89–113. [Google Scholar]
- Legrand, P.H.; Guillaumin, J.J. Armillaria species in the forest ecosystems of the Auvergne (central France). Acta Oecol. 1993, 14, 389–403. [Google Scholar]
- Bruhn, J.N.; Wetteroff, J.J., Jr.; Mihail, J.D.; Kabrick, J.M.; Pickens, J.B. Distribution of Armillaria species in upland Ozark Mountain forests with respect to site, overstory species composition and oak decline. For. Pathol. 2000, 30, 43–60. [Google Scholar] [CrossRef]
- Thomidis, T.; Exadaktylou, E. Effectiveness of cyproconazole to control Armillaria root rot of apple, walnut and kiwifruit. Crop Prot. 2012, 36, 49–51. [Google Scholar] [CrossRef]
- Beckman, T.G.; Okie, W.R.; Nyczepir, A.P.; Pusey, P.L.; Reilly, C.C. Relative susceptibility of peach and plum germplasm to Armillaria root rot. HortScience 1998, 33, 1062–1065. [Google Scholar] [CrossRef] [Green Version]
- Guillaumin, J.-J.; Mohammed, C.; Anselmi, N.; Courtecuisse, R.; Gregory, S.C.; Holdenrieder, O.; Intini, M.; Lung, B.; Marxmüller, H.; Morrison, D.; et al. Geographical distribution and ecology of the Armillaria species in western Europe. Eur. J. For. Pathol. 1993, 23, 321–341. [Google Scholar] [CrossRef]
- Baumgartner, K.; Rizzo, D.M. Distribution of Armillaria species in California. Mycologia 2001, 93, 821–830. [Google Scholar] [CrossRef]
- Nelson, E.V.; Fairweather, M.L.; Ashiglar, S.M.; Hanna, J.W.; Klopfenstein, N.B. First report of the Armillaria root disease pathogen, Armillaria gallica, on Douglas-fir (Pseudotsuga menziesii) in Arizona. Plant Dis. 2013, 97, 1658. [Google Scholar] [CrossRef]
- Klopfenstein, N.B.; Hanna, J.W.; Cannon, P.G.; Medel-Ortiz, R.; Alvarado-Rosales, D.; Lorea-Hernández, F.; Elías-Román, R.D.; Kim, M.-S. First report of the Armillaria root-disease pathogen, Armillaria gallica, associated with several woody hosts in three states of Mexico. Plant Dis. 2014, 98, 1280. [Google Scholar] [CrossRef]
- Kim, M.S.; Hanna, J.W.; Klopfenstein, N.B. First report of an Armillaria root disease pathogen, Armillaria gallica, associated with several new hosts in Hawaii. Plant Dis. 2010, 94, 1503. [Google Scholar] [CrossRef]
- Duarte-Mata, E.; Elias, R.; Hanna, J.W.; Klopfenstein, N.B.; Kim, M.-S. First report of the Armillaria root-disease pathogen, Armillaria gallica, associated with several woody hosts in three states of central Mexico (Guanajuato, Jalisco, and Michoacan). Plant Dis. 2020, in press. [Google Scholar] [CrossRef]
- Volk, T.J.; Burdsall, H.H.; Banik, M.T. Armillaria nabsnona, a new species from western North America. Mycologia 1996, 88, 484–491. [Google Scholar] [CrossRef]
- Ota, Y.; Sotome, K.; Hasegawa, E. Seven Armillaria species identified from Hokkaido Island, northern Japan. Mycoscience 2009, 50, 442–447. [Google Scholar] [CrossRef]
- Ota, Y.; Matsushita, N.; Nagasawa, E.; Terashita, T.; Fukuda, K.; Suzuki, K. Biological species of Armillaria in Japan. Plant Dis. 1998, 82, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Brazee, N.J.; Ortiz-Santana, B.; Banik, M.T.; Lindner, D.L. Armillaria altimontana, a new species from the western interior of North America. Mycologia 2012, 104, 1200–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonín, V.; Jankovsky, L.; Lochman, J.; Tomsovsky, M. Armillaria socialis—Morphological-anatomical and ecological characteristics, pathology, distribution in the Czech Republic and Europe and remarks on its genetic variation. Czech Mycol. 2006, 58, 209. [Google Scholar] [CrossRef] [Green Version]
- Schnabel, G.; Ash, J.S.; Bryson, P.K. Identification and characterization of Armillaria tabescens from the southeastern United States. Mycol. Res. 2005, 109, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Sipos, G.; Anderson, J.B.; Nagy, L.G. Armillaria. Curr. Biol. 2018, 28, R297–R298. [Google Scholar] [CrossRef] [Green Version]
- Sahu, N.; Merényi, Z.; Bálint, B.; Kiss, B.; Sipos, G.; Owens, R.A.; Nagy, L.G. Hallmarks of Basidiomycete soft- and white-rot in wood-decay - Omics data of two Armillaria species. Microorganisms 2021, 9, 149. [Google Scholar] [CrossRef]
- Heinzelmann, R.; Rigling, D.; Sipos, G.; Münsterkötter, M.; Croll, D. Chromosomal assembly and analyses of genome-wide recombination rates in the forest pathogenic fungus Armillaria ostoyae. Heredity 2020, 124, 699–713. [Google Scholar] [CrossRef]
- Collins, C.; Keane, T.M.; Turner, D.J.; O’Keeffe, G.; Fitzpatrick, D.A.; Doyle, S. Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: Toward a new infection model system. J. Proteome Res. 2013, 12, 2552–2570. [Google Scholar] [CrossRef]
- Wingfield, B.D.; Ambler, J.M.; Coetzee, M.P.A.; de Beer, Z.W.; Duong, T.A.; Joubert, F.; Hammerbacher, A.; McTaggart, A.R.; Naidoo, K.; Nguyen, H.D.T.; et al. IMA Genome-F 6: Draft genome sequences of Armillaria fuscipes, Ceratocystiopsis minuta, Ceratocystis adiposa, Endoconidiophora laricicola, E. polonica and Penicillium freii DAOMC 242723. IMA Fungus 2016, 7, 217–227. [Google Scholar] [CrossRef]
- Anderson, J.B.; Bruhn, J.N.; Kasimer, D.; Wang, H.; Rodrigue, N.; Smith, M.L. Clonal evolution and genome stability in a 2500-year-old fungal individual. Proc. Biol. Sci. 2018, 285, 20182233. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Tian, M.; Wang, W.; Li, G.; Lu, X.; Cai, G.; Yang, H.; Du, G.; Huang, L. Draft genomic sequence of Armillaria gallica 012m: Insights into its symbiotic relationship with Gastrodia elata. Braz. J. Microbiol. 2020, 51, 1539–1552. [Google Scholar] [CrossRef] [PubMed]
- Devkota, P.; Hammerschmidt, R. The infection process of Armillaria mellea and Armillaria solidipes. Physiol. Mol. Plant Pathol. 2020, 112, 101543. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Whiting, E.C.; Elkins, R.B. Spatial distribution of Armillaria mellea in pear orchards. Plant Dis. 1998, 82, 1226–1231. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Lung-Escarmant, B.; Dutech, C. Genetic structure of an expanding Armillaria root rot fungus (Armillaria ostoyae) population in a managed pine forest in southwestern France. Mol. Ecol. 2008, 17, 3366–3378. [Google Scholar] [CrossRef] [PubMed]
- Labbé, F.; Fontaine, M.C.; Robin, C.; Dutech, C. Genetic signatures of variation in population size in a native fungal pathogen after the recent massive plantation of its host tree. Heredity 2017, 119, 402–410. [Google Scholar] [CrossRef]
- Lehtijärvi, A.; Doğmuş-Lehtijärvi, H.T.; Aday, A.G. Armillaria ostoyae associated with dying 60-year-old Scots pines in northern Turkey. For. Pathol. 2012, 42, 267–269. [Google Scholar] [CrossRef]
- Smith, M.L.; Bruhn, J.N.; Anderson, J.B. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature 1992, 356, 428–431. [Google Scholar] [CrossRef]
- Solla, A.; Tomlinson, F.; Woodward, S. Penetration of Picea sitchensis root bark by Armillaria mellea, Armillaria ostoyae and Heterobasidion annosum. For. Pathol. 2002, 32, 55–70. [Google Scholar] [CrossRef]
- Cairney, J.W.; Jennings, D.H.; Ratcliffe, R.G.; Southon, T.E. The physiology of basidiomycete linear organs II. Phosphate uptake by rhizomorphs of Armillaria mellea. New Phytol. 1988, 109, 327–333. [Google Scholar] [CrossRef]
- Pareek, M.; Cole, L.; Ashford, A.E. Variations in structure of aerial and submerged rhizomorphs of Armillaria luteobubalina indicate that they may be organs of absorption. Mycol. Res. 2001, 105, 1377–1387. [Google Scholar] [CrossRef]
- Yafetto, L.; Davis, D.J.; Money, N.P. Biomechanics of invasive growth by Armillaria rhizomorphs. Fungal Genet. Biol. 2009, 46, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Shaw, C.G., III. Basidiospores of Armillaria mellea survive an Alaskan winter. Plant Dis. 1981, 65, 972–974. [Google Scholar] [CrossRef]
- Marçais, B.; Breda, N. Role of an opportunistic pathogen in the decline of stressed oak trees. J. Ecol. 2006, 94, 1214–1223. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.J. Rhizomorph growth habit, saprophytic ability and virulence of 15 Armillaria species. For. Pathol. 2004, 34, 15–26. [Google Scholar] [CrossRef]
- Guo, T.; Wang, H.C.; Xue, W.Q.; Zhao, J.; Yang, Z.L. Phylogenetic analyses of Armillaria reveal at least 15 phylogenetic lineages in China, seven of which are associated with cultivated Gastrodia elata. PLoS ONE 2016, 11, e0154794. [Google Scholar] [CrossRef] [Green Version]
- Mihail, J.D.; Bruhn, J.N. Foraging behaviour of Armillaria rhizomorph systems. Mycol. Res. 2005, 109, 1195–1207. [Google Scholar] [CrossRef]
- Pareek, M.; Allaway, W.G.; Ashford, A.E. Armillaria luteobubalina mycelium develops air pores that conduct oxygen to rhizomorph clusters. Mycol. Res. 2006, 110, 38–50. [Google Scholar] [CrossRef]
- Gonthier, P. Controlling root and butt rot diseases in alpine European forests. In Management of Fungal Plant Pathogens; Arya, A., Perelló, A., Eds.; CAB International: Cambridge, MA, USA, 2010; Volume 26, pp. 345–361. [Google Scholar] [CrossRef]
- Cleary, M.R.; Arhipova, N.; Morrison, D.J.; Thomsen, I.M.; Sturrock, R.N.; Vasaitis, R.; Gaitnieks, T.; Stenlid, J. Stump removal to control root disease in Canada and Scandinavia: A synthesis of results from long-term trials. For. Ecol. Manag. 2013, 290, 5–14. [Google Scholar] [CrossRef]
- Lung-Escarmant, B.; Guyon, D. Temporal and spatial dynamics of primary and secondary infection by Armillaria ostoyae in a Pinus pinaster plantation. Phytopathology 2004, 94, 125–131. [Google Scholar] [CrossRef]
- Heinzelmann, R.; Prospero, S.; Rigling, D. Frequent diploidisation of haploid Armillaria ostoyae strains in an outdoor inoculation experiment. Fungal Biol. 2018, 122, 147–155. [Google Scholar] [CrossRef]
- Peabody, R.B.; Peabody, D.C.; Tyrrell, M.G.; Edenburn-MacQueen, E.; Howdy, R.P.; Semelrath, K.M. Haploid vegetative mycelia of Armillaria gallica show among-cell-line variation for growth and phenotypic plasticity. Mycologia 2005, 97, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, M.G.; Peabody, D.C.; Peabody, R.B.; James-Pederson, M.; Hirst, R.G.; Allan-Perkins, E.; Bickford, H.; Shafrir, A.; Doiron, R.J.; Churchill, A.C.; et al. Mosaic fungal individuals have the potential to evolve within a single generation. Sci. Rep. 2020, 10, 17625. [Google Scholar] [CrossRef]
- Legrand, P.; Ghahari, S.; Guillaumin, J.J. Occurrence of genets of Armillaria spp. in four mountain forests in central France: The colonization strategy of Armillaria ostoyae. New Phytol. 1996, 133, 321–332. [Google Scholar] [CrossRef]
- Dutech, C.; Labbé, F.; Capdevielle, X.; Lung-Escarmant, B. Genetic analysis reveals efficient sexual spore dispersal at a fine spatial scale in Armillaria ostoyae, the causal agent of root-rot disease in conifers. Fungal Biol. 2017, 121, 550–560. [Google Scholar] [CrossRef]
- Bendel, M.; Kienast, F.; Rigling, D.; Bugmann, H. Impact of root-rot pathogens on forest succession in unmanaged Pinus mugo stands in the central Alps. Can. J. For. Res. 2006, 36, 2666–2674. [Google Scholar] [CrossRef]
- Cleary, M.R.; van der Kamp, B.J.; Morrison, D.J. Effects of wounding and fungal infection with Armillaria ostoyae in three conifer species. I. Host response to abiotic wounding in non-infected roots. For. Pathol. 2012, 42, 100–108. [Google Scholar] [CrossRef]
- Wong, J.W.-H.; Plett, K.L.; Natera, S.H.A.; Roessner, U.; Anderson, I.C.; Plett, J.M. Comparative metabolomics implicates threitol as a fungal signal supporting colonization of Armillaria luteobubalina on eucalypt roots. Plant Cell Environ. 2020, 43, 374–386. [Google Scholar] [CrossRef]
- Schwarze, F.W.M.R.; Baum, S.; Fink, S. Resistance of fibre regions in wood of Acer pseudoplatanus degraded by Armillaria mellea. Mycol. Res. 2000, 104, 126–132. [Google Scholar] [CrossRef]
- Schwarze, F.W.M.R. Wood decay under the microscope. Fungal Biol. Rev. 2007, 21, 133–170. [Google Scholar] [CrossRef]
- Baumgartner, K.; Warnock, A.E. A soil inoculant inhibits Armillaria mellea in vitro and improves productivity of grapevines with root disease. Plant Dis. 2006, 90, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Pertot, I.; Gobbin, D.; De Luca, F.; Prodorutti, D. Methods of assessing the incidence of Armillaria root rot across viticultural areas and the pathogen’s genetic diversity and spatial–temporal pattern in northern Italy. Crop Prot. 2008, 27, 1061–1070. [Google Scholar] [CrossRef]
- Morrison, D.J.; Williams, R.E.; Whitney, R.D. Infection, disease development, diagnosis, and detection. In Armillaria Root Disease; Agriculture Handbook No. 691; Show, C.G., III, Kile, G.A., Eds.; United States Department of Agriculture, Forest Service: Washington, DC, USA, 1991; pp. 62–75. [Google Scholar]
- Bendel, M.; Rigling, D. Signs and symptoms associated with Heterobasidion annosum and Armillaria ostoyae infection in dead and dying mountain pine (Pinus mugo ssp. uncinata). For. Pathol. 2008, 38, 61–72. [Google Scholar] [CrossRef]
- Holuša, J.; Lubojacký, J.; Čurn, V.; Tonka, T.; Lukášová, K.; Horák, J. Combined effects of drought stress and Armillaria infection on tree mortality in Norway spruce plantations. For. Ecol. Manag. 2018, 427, 434–445. [Google Scholar] [CrossRef]
- Worrall, J.J.; Sullivan, K.F.; Harrington, T.C.; Steimel, J.P. Incidence, host relations and population structure of Armillaria ostoyae in Colorado campgrounds. For. Ecol. Manag. 2004, 192, 191–206. [Google Scholar] [CrossRef]
- Lundquist, J.E. A method of estimating direct and indirect effects of Armillaria root disease and other small-scale forest disturbances on canopy gap size. For. Sci. 2000, 46, 356–362. [Google Scholar] [CrossRef]
- Mallett, K.I.; Volney, W.J. The effect of Armillaria root disease on lodgepole pine tree growth. Can. J. For. Res. 1999, 29, 252–259. [Google Scholar] [CrossRef]
- Westwood, A.R.; Conciatori, F.; Tardif, J.C.; Knowles, K. Effects of Armillaria root disease on the growth of Picea mariana trees in the boreal plains of central Canada. For. Ecol. Manag. 2012, 266, 1–10. [Google Scholar] [CrossRef]
- Rosso, P.; Hansen, E. Tree vigour and the susceptibility of Douglas fir to Armillaria root disease. Eur. J. For. Pathol. 1998, 28, 43–52. [Google Scholar] [CrossRef]
- Lee, C.A.; Dey, D.C.; Muzika, R.M. Oak stump-sprout vigor and Armillaria infection after clearcutting in southeastern Missouri, USA. For. Ecol. Manag. 2016, 374, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Kovalchuk, A.; Keriö, S.; Oghenekaro, A.O.; Jaber, E.; Raffaello, T.; Asiegbu, F.O. Antimicrobial defenses and resistance in forest trees: Challenges and perspectives in a genomic era. Annu. Rev. Phytopathol. 2013, 51, 221–244. [Google Scholar] [CrossRef]
- Barrett, L.G.; Kniskern, J.M.; Bodenhausen, N.; Zhang, W.; Bergelson, J. Continua of specificity and virulence in plant host-pathogen interactions: Causes and consequences. New Phytol. 2009, 183, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.M.; Morrison, D.J.; Jensen, G.D. Necrophylactic periderm formation in the roots of western larch and Douglas-fir trees infected with Armillaria ostoyae. II. The response to the pathogen. For. Pathol. 2004, 34, 119–129. [Google Scholar] [CrossRef]
- Brazee, N.J.; Wick, R.L.; Wargo, P.M. Effects of hydrolyzable tannins on in vitro growth of Armillaria calvescens and A. gallica. Plant Dis. 2011, 95, 1255–1262. [Google Scholar] [CrossRef]
- Wargo, P.M.; Harrington, T.C. Host stress and susceptibility. In Armillaria Root Disease; Agriculture Handbooks AH691; Shaw, C.G., III, Kile, G.A., Eds.; USDA: Washington, DC, USA, 1991; pp. 88–101. [Google Scholar]
- Kubiak, K.; Żółciak, A.; Damszel, M.; Lech, P.; Sierota, Z. Armillaria pathogenesis under climate changes. Forests 2017, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Ayres, M.P.; Lombardero, M.J. Assessing the consequences of global change for forest disturbance from herbivores and pathogens. Sci. Total Environ. 2000, 262, 263–286. [Google Scholar] [CrossRef]
- Kliejunas, J.T.; Geils, B.W.; Glaeser, J.M.; Goheen, E.M.; Hennon, P.; Kim, M.S.; Kope, H.; Stone, J.L.; Sturrock, R.; Frankel, S.J. Climate and Forest Diseases of Western North America: A Literature Review; General Technical Report PSW-GTR-225; United States Department of Agriculture, Forest Service, Pacific Southwest Research Station: Washington, DC, USA, 2009.
- Pearce, M.H.; Malajczuk, N. Factors affecting growth of Armillaria luteobubalina rhizomorphs in soil. Mycol. Res. 1990, 94, 38–48. [Google Scholar] [CrossRef]
- Husson, C.; Cael, O.; Grandjean, J.P.; Nageleisen, L.M.; Marcais, B. Occurrence of Hymenoscyphus pseudoalbidus on infected ash logs. Plant Pathol. 2012, 61, 889–895. [Google Scholar] [CrossRef]
- Kwaśna, H.; Łakomy, P.; Mallett, K. Reaction of Armillaria ostoyae to forest soil microfungi. For. Pathol. 2004, 34, 147–162. [Google Scholar] [CrossRef]
- Kwaśna, H.; Lakomy, P. Stimulation of Armillaria ostoyae vegetative growth by tryptophol and rhizomorph produced by Zygorhynchus moelleri. Eur. J. Plant Pathol. 1998, 28, 53–61. [Google Scholar] [CrossRef]
- Kwaśna, H. Fungi in the rhizosphere of common oak and its stumps and their possible effect on infection by Armillaria. Appl. Soil Ecol. 2001, 17, 215–227. [Google Scholar] [CrossRef]
- Kwaśna, H. Changes in microfungal communities in roots of Quercus robur stumps and their possible effect on colonization by Armillaria. J. Phytopathol. 2002, 150, 403–411. [Google Scholar] [CrossRef]
- Kwaśna, H. The effect of felling on the occurrence of microfungi stimulatory to Armillaria rhizomorph formation in thin roots of Quercus robur. J. Phytopathol. 2003, 151, 185–189. [Google Scholar] [CrossRef]
- Baker, W.L. Effect of gypsy moth defoliation on certain forest trees. J. For. 1941, 39, 1017–1022. [Google Scholar] [CrossRef]
- Kegg, J.D. The impact of gypsy moth: Repeated defoliation of oak in New Jersey. J. For. 1971, 69, 852–854. [Google Scholar]
- Karasevicz, D.; Merrill, W. Succession of biodeterioration fungi in oaks killed following gypsy moth defoliation in Pennsylvania. Annual Meeting of the American Phytopathological Society, Potomac Division, 2–4. April 1986. Phytopathology 1986, 76, 564. [Google Scholar]
- Twery, M.J.; Mason, G.N.; Wargo, P.M.; Gottschalk, K.W. Abundance and distribution of rhizomorphs of Armillaria spp. in defoliated mixed oak stands in western Maryland. Can. J. For. Res. 1990, 20, 674–678. [Google Scholar] [CrossRef]
- Burrill, E.A.; Worrall, J.J.; Wargo, P.M.; Stehman, S.V. Effects of defoliation and cutting in eastern oak forests on Armillaria spp. and a competitor, Megacollybia platyphylla. Can. J. For. Res. 1999, 29, 347–355. [Google Scholar] [CrossRef]
- Stilwell, M.A.; Kelly, D.J. Fungous deterioration of balsam fir killed by spruce budworm in Northwestern New Brunswick. For. Chron. 1964, 40, 482–487. [Google Scholar] [CrossRef] [Green Version]
- Sterner, I.E. Butt decay in balsam fir defoliated by the spruce budworm. Can. Dept. Fish. For. Bi. Mon. Res. Notes 1970, 26, 38–39. [Google Scholar]
- Raske, A.G.; Sutton, W.J. Decline and Mortality of Black Spruce Caused by Spruce Budworm Defoliation and Secondary Organisms; Inf. Rep. N-X-236; Newfoundland Forestry Centre, Canadian Forestry Centre: Edmonton, AB, Canada, 1986; p. 29. [Google Scholar]
- Filip, G.M. Interactions among root diseases and agents of defoliation. In Proceedings of the 7th International Conference on Root and Butt Rots, Vernon and Victoria, BC, Canada, 9–16 August 1988; Morrison, D.J., Ed.; International Union of Forestry Research Organizations: Vernon and Victoria, BC, Canada, 1989; pp. 149–155. [Google Scholar]
- Houston, D.R.; Kuntz, J.E. Studies of the maple blight. Part III. Pathogens associated with maple blight. Univ. Wiscons. Res. Bull. 1964, 250, 59–79. [Google Scholar]
- Staley, J.M. Decline and mortality of red and scarlet oaks. For. Sci. 1965, 11, 2–17. [Google Scholar] [CrossRef]
- Austarå, Ø. Diametertilvekst og Tredødelighet Etter Masseangrep av Liten Granbarvikler (Diameter Growth and Tree Mortality of Norway Spruce Following Mass Attacks by Epinotia Nanana); NISK: Ås, Norway, 1984. [Google Scholar]
- Madziara-Borusiewicz, K.; Strzelecka, H. Conditions of spruce (Picea excelsa Lk.) infestation by the engraver beetle (Ips typographus L.) in mountains of Poland. Zeitschr. Angew. Entomol. 1977, 83, 409–415. [Google Scholar] [CrossRef]
- Jakuš, R. Bark beetle (Coleoptera, Scolytidae) outbreak and system of IPM measures in an area affected by intensive forest decline connected with honey fungus (Armillaria sp.). Anz. Schädlingskunde J. Pest Sci. 2001, 74, 46–51. [Google Scholar] [CrossRef]
- Grodzki, W. Spatio-temporal patterns of the Norway spruce decline in the Beskid Śląski and Żywiecki (Western Carpathians) in southern Poland. J. For. Sci. 2007, 53, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Jankovský, L.; Cudlín, P.; Moravec, I. Root decays as a potential predisposition factor of a bark beetle disaster in the Šumava Mts. J. For. Sci. 2003, 49, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Hertert, H.D.; Miller, D.L.; Partridge, A.D. Interaction of bark beetles (Coleoptera: Scolytidae) and root-rot pathogens in grand fir in Northern Idaho. Can. Entomol. 1975, 107, 899–904. [Google Scholar] [CrossRef]
- Miller, D.L.; Partridge, A.D. Root-rot indicators in grand fir. Plant Dis. Rep. 1974, 58, 275–276. [Google Scholar]
- James, R.L.; Goheen, D.J. Conifer mortality associated with root disease and insects in Colorado. Plant Dis. 1981, 65, 506–507. [Google Scholar] [CrossRef]
- Lane, B.B.; Goheen, D.J. Incidence of root disease in bark beetle-infested eastern Oregon and Washington true firs. Plant Dis. Rep. 1979, 63, 262–266. [Google Scholar]
- Filip, G.M. Forest health decline in Central Oregon: A 13-year case study. Northwest Sci. 1994, 68, 233–240. [Google Scholar]
- Whitney, R.D. Root wounds and associated root rots of white spruce. For. Chron. 1961, 37, 401–411. [Google Scholar] [CrossRef]
- Warren, G.L.; Singh, P. Hylobius Weevils and Armillaria Root Rot in a Coniferous Plantation in Newfoundland; Nfld. Bi-monthly Research Notes; Canada Fisheries and Forestry, Canadian Forestry Service: St. John, NL, Canada, 1970; Volume 26, p. 55. [Google Scholar]
- Kulhavy, D.L.; Partridge, A.D.; Stark, R.W. Root diseases and blister rust associated with bark beetles (Coleoptera: Scolytidae) in Western white pine in Idaho. Environ. Entomol. 1984, 813–817. [Google Scholar] [CrossRef]
- Tkacz, B.M.; Schmitz, R.F. Association of an Endemic Mountain Pine Beetle Population with Lodgepole Pine Infected by Armillaria Root Disease in Utah; Research Note INT-353; United States Department of Agriculture, Forest Service: Washington, DC, USA, 1986.
- Tunnock, S.; Denton, R.E.; Carlson, C.C. Larch Casebearer and Other Factors Involved with Deterioration of Western Larch Stands in Northern Idaho; Forest Service Research Paper INT-68; US Department of Agriculture: Ogden, UT, USA, 1969; p. 10.
- Dunbar, D.M.; Stephens, G.R. Association of two-lined chestnut borer and shoestring fungus with mortality of defoliated oaks in Connecticut. For. Sci. 1975, 21, 169–174. [Google Scholar]
- Kula, E.; Zabecki, W. Cambioxylophagous niche on spruce-trees affected by the stress of root fungal pathogens. J. For. Sci. UZPI 1999, 45, 348–357. [Google Scholar]
- Hudak, J.; Singh, P. Incidence of Armillaria root rot in balsam fir infested by balsam woolly aphid. Can. Plant Dis. Serv. 1970, 50, 99–101. [Google Scholar]
- Hudak, J.; Wells, R.E. Armillaria root rot in aphid-damaged balsam fir in Newfoundland. For. Chron. 1974, 50, 74–76. [Google Scholar] [CrossRef] [Green Version]
- Wargo, P.M.; Houston, D.R. Infection of defoliated sugar maple trees by Armillaria mellea. Phytopathology 1974, 64, 817–822. [Google Scholar] [CrossRef]
- Sierota, Z.; Grodzki, W. Picea abies–Armillaria–Ips: A strategy or coincidence? Forests 2020, 11, 1023. [Google Scholar] [CrossRef]
- Chapman, W.K.; Schellenberg, B.; Newsome, T.A. Assessment of Armillaria root disease infection in stands in south-central British Columbia with varying levels of overstory retention, with and without pushover logging. Can. J. For. Res. 2011, 41, 1598–1605. [Google Scholar] [CrossRef]
- Baietto, M.; Wilson, A.D. Relative in vitro wood decay resistance of sapwood from landscape trees of southern temperate regions. HortScience 2010, 45, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Cruickshank, M.G.; Jaquish, B.; Nemec, A.F. Resistance of half-sib interior Douglas-fir families to Armillaria ostoyae in British Columbia following artificial inoculation. Can. J. For. Res. 2010, 40, 155–166. [Google Scholar] [CrossRef]
- Cruickshank, M.G.; Jaquish, B. Resistance and tolerance in juvenile interior Douglas-fir trees Pseudotsuga menziesii var. glauca artificially inoculated with Armillaria ostoyae. For. Pathol. 2014, 44, 362–371. [Google Scholar] [CrossRef]
- Metaliaj, R.; Sicoli, G.; Luisi, N. Pathogenicity of Armillaria isolates inoculated on five Quercus species at different watering regimes. Phytopathol. Mediterr. 2006, 45, 3–9. [Google Scholar] [CrossRef]
- Elkins, R.B.; Rizzo, D.M.; Whiting, E.C. Biology and management of Armillaria root disease in pear in California. Acta Horticult. 1997, 475, 453–458. [Google Scholar] [CrossRef]
- Redfern, D.B.; Filip, G.M. Inoculum and infection. In Armillaria Root Disease; Agriculture Handbook No. 691; Show, C.G., III, Kile, G.A., Eds.; United States Department of Agriculture, Forest Service: Washington, DC, USA, 1991; pp. 48–61. [Google Scholar]
- West, J.S. Chemical control of Armillaria. In Armillaria Root Rot: Biology and Control of Honey Fungus; Fox, R.T.V., Ed.; Intercept: Andover, Hampshire, UK, 2000; pp. 173–182. [Google Scholar]
- Pronos, J.; Patton, R.F. Penetration and colonization of oak roots by Armillaria mellea in Wisconsin. Eur. J. For. Pathol. 1978, 8, 259–267. [Google Scholar] [CrossRef]
- Vasaitis, R.; Stenlid, J.; Thomsen, I.M.; Barklund, P.; Dahlberg, A. Stump removal to control root rot in forest stands. A literature study. Silva Fenn. 2008, 42, 457. [Google Scholar] [CrossRef] [Green Version]
- Sturrock, R.N. Management of Root Diseases by Stumping and Push-Falling; Pacific Forestry Centre, Canadian Forest Service: Ottawa, QC, Canada, 2000. [Google Scholar]
- Shaw, C.G., III; Omdal, D.W.; Ramsey-Kroll, A.; Roth, L.F. Inoculum reduction measures to manage Armillaria root disease in a severely infected stand of ponderosa pine in south-central Washington: 35-year results. West. J. Appl. For. 2012, 27, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Hagle, S.K.; Shaw, C.G., III. Avoiding and reducing losses from Armillaria root disease. In Armillaria Root Disease; Agriculture Handbook No. 691; Show, C.G., III, Kile, G.A., Eds.; United States Department of Agriculture, Forest Service: Washington, DC, USA, 1991; pp. 157–173. [Google Scholar]
- Morrison, D.J.; Cruickshank, M.G.; Lalumière, A. Control of laminated and Armillaria root diseases by stump removal and tree species mixtures: Amount and cause of mortality and impact on yield after 40 years. For. Ecol. Manag. 2014, 319, 75–98. [Google Scholar] [CrossRef]
- Shaw, C.G., III; Roth, L.F. Control of Armillaria root rot in managed coniferous forests 1: A literature review. Eur. J. For. Pathol. 1978, 8, 163–174. [Google Scholar] [CrossRef]
- Schüti, P. Control of root and butt rots: Limits and prospects. Eur. J. For. Pathol. 1985, 15, 357–363. [Google Scholar] [CrossRef]
- Fox, R.T.V. (Ed.) Armillaria root rot: Biology and control of honey fungus. N. Z. J. For. Sci. 2000, 31, 150–152. [Google Scholar] [CrossRef]
- Self, N.M.; MacKenzie, M.A. Intensive site-preparation to control Armillaria root disease in second-rotation Pinus radiata. N. Z. J. For. Sci. 1995, 25, 111–116. [Google Scholar]
- Otieno, W.; Jeger, M.; Termorshuizen, A. Effect of infesting soil with Trichoderma harzianum and amendment with coffee pulp on survival of Armillaria. Biol. Control 2003, 26, 293–301. [Google Scholar] [CrossRef]
- Otieno, W.; Termorshuizen, A.; Jeger, M.; Othieno, C.O. Efficacy of soil solarization, Trichoderma harzianum, and coffee pulp amendment against Armillaria sp. Crop Prot. 2003, 22, 325–331. [Google Scholar] [CrossRef]
- Robinson, R.M.; Smith, R.H. Fumigation of regrowth karri stumps with metham-sodium to control Armillaria luteobubalina. Austral. For. 2001, 64, 209–215. [Google Scholar] [CrossRef]
- Raziq, F. 2000. Biological and integrated control of Armillaria root rot. In Armillaria Root Rot: Biology and Control of Honey Fungus; Fox, R.T.V., Ed.; Intercept: Andover, UK, 2000; pp. 183–201. [Google Scholar]
- Singh, N.; Pandey, P.; Dubey, R.C.; Maheshwari, D.K. Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii (Sarg.) by rhizosphere competent Bacillus subtilis BN1. World J. Microbiol. Biotechnol. 2008, 24, 1669. [Google Scholar] [CrossRef]
- Whipps, J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001, 52, 487–511. [Google Scholar] [CrossRef]
- DeLong, R.L.; Lewis, K.J.; Simard, S.W.; Gibson, S. Fluorescent pseudomonad population sizes baited from soils under pure birch, pure Douglas-fir, and mixed forest stands and their antagonism toward Armillaria ostoyae in vitro. Can. J. For. Res. 2002, 32, 2146–2159. [Google Scholar] [CrossRef]
- Dumas, M.T. Inhibition of Armillaria by bacteria isolated from soils of the boreal mixedwood forest of Ontario. Eur. J. For. Pathol. 1992, 22, 11–18. [Google Scholar] [CrossRef]
- Zagryadskaya, Y.A.; Lysak, L.V.; Chernov, I.Y. Bacterial communities in the fruit bodies of ground basidiomycetes. Euras. Soil Sci. 2015, 48, 620–626. [Google Scholar] [CrossRef]
- Mesanza, N.; Iturritxa, E.; Patten, C.L. Native rhizobacteria as biocontrol agents of Heterobasidion annosum s.s. and Armillaria mellea infection of Pinus radiata. Biol. Control 2016, 101, 8–16. [Google Scholar] [CrossRef]
- de Vasconcellos, R.L.; Cardoso, E.J. Rhizospheric streptomycetes as potential biocontrol agents of Fusarium and Armillaria pine rot and as PGPR for Pinus taeda. Biocontrol 2009, 54, 807. [Google Scholar] [CrossRef]
- Maier, A.; Riedlinger, J.; Fiedler, H.P.; Hampp, R. Actinomycetales bacteria from a spruce stand: Characterization and effects on growth of root symbiotic and plant parasitic soil fungi in dual culture. Mycol. Progr. 2004, 3, 129–136. [Google Scholar] [CrossRef]
- Harman, G.E.; Kubicek, C.P. (Eds.) Trichoderma and Gliocladium, Volume 2: Enzymes, Biological Control and Commercial Applications; Taylor and Francis: London, UK, 1998. [Google Scholar] [CrossRef]
- Verma, M.; Brar, S.K.; Tyagi, R.D.; Surampalli, R.Y.; Valero, J.R. Antagonistic fungi, Trichoderma spp.: Panoply of biological control. Biochem. Eng. J. 2007, 37, 1–20. [Google Scholar] [CrossRef]
- Perazzolli, M.; Antonielli, L.; Storari, M.; Puopolo, G.; Pancher, M.; Giovannini, O.; Pindo, M.; Pertot, I. Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl. Environ. Microb. 2014, 80, 3585–3596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrini, A.; Corneo, P.E.; Camin, F.; Ziller, L.; Tosi, S.; Pertot, I. Studying trophic interactions between a plant pathogen and two different antagonistic microorganisms using a 13C-labeled compound and isotope ratio mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 510–516. [Google Scholar] [CrossRef]
- Longa, C.M.; Pertot, I.; Tosi, S. Ecophysiological requirements and survival of a Trichoderma atroviride isolate with biocontrol potential. J. Basic Microb. 2008, 48, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, A.; Prodorutti, D.; Pellegrini, C.; Paternoster, T.; Leoni, V.; Pertot, I. Use of Trichoderma atroviride SC1 inoculated barks to control Armillaria root rot in highbush blueberry orchards. IOBC/WPRS Bull. 2009, 43, 259–262. [Google Scholar]
- Elad, Y.; Barak, R.; Chet, I.; Henis, Y. Ultrastructural studies of the interaction between Trichoderma spp. and plant pathogenic fungi. J. Phytopathol. 1983, 107, 168–175. [Google Scholar] [CrossRef]
- Dumas, M.T.; Boyonoski, N.W. Scanning electron microscopy of mycoparasitism of Armillaria rhizomorphs by species of Trichoderma. Eur. J. For. Pathol. 1992, 22, 379–383. [Google Scholar] [CrossRef]
- Onsando, J.M.; Waudo, S.W. Interaction between Trichoderma species and Armillaria root rot fungus of tea in Kenya. Int. J. Pest Manag. 1994, 40, 69–74. [Google Scholar] [CrossRef]
- Reaves, J.L.; Shaw, C.G.; Mayfield, J.E. The effect of Trichoderma spp. isolated from burned and non-burned forest soils on the growth and development of Armillaria ostoyae in culture. Northwest Sci. 1990, 6, 39–44. [Google Scholar]
- Tarus, P.K.; Lang’at-Thoruwa, C.C.; Wanyonyi, A.W.; Chhabra, S.C. Bioactive metabolites from Trichoderma harzianum and Trichoderma longibrachiatum. Bull. Chem. Soc. Ethiopia 2003, 17, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Pellegrini, A.; Corneo, P.E.; Camin, F.; Ziller, L.; Tosi, S.; Pertot, I. Isotope ratio mass spectrometry identifies soil microbial biocontrol agents having trophic relations with the plant pathogen Armillaria mellea. Appl. Soil Ecol. 2013, 64, 142–151. [Google Scholar] [CrossRef]
- Raziq, F.; Fox, R.T. The effect of carrier substrate, dose rate and time of application on biocontrol efficacy of fungal antagonists against Armillaria root rot of strawberry plants. Biol. Agric. Horticult. 2004, 22, 157–172. [Google Scholar] [CrossRef]
- Savazzini, F.; Longa, C.M.; Pertot, I. Impact of the biocontrol agent Trichoderma atroviride SC1 on soil microbial communities of a vineyard in northern Italy. Soil Biol. Biochem. 2009, 41, 1457–1465. [Google Scholar] [CrossRef]
- Percival, G.C.; Smiley, E.T.; Fox, R.T. Root collar excavation with Trichoderma inoculations as a potential management strategy for honey fungus (Armillaria mellea). Arboricult. J. 2011, 33, 267–280. [Google Scholar] [CrossRef]
- Chen, L.; Bóka, B.; Kedves, O.; Nagy, V.D.; Szűcs, A.; Champramary, S.; Roszik, R.; Patocskai, Z.; Münsterkötter, M.; Huynh, T.; et al. Towards the biological control of devastating forest pathogens from the genus Armillaria. Forests 2019, 10, 1013. [Google Scholar] [CrossRef] [Green Version]
- Rees, H.J.; Bashir, N.; Drakulic, J.; Cromey, M.G.; Bailey, A.M.; Foster, G.D. Identification of native endophytic Trichoderma spp. for investigation of in vitro antagonism towards Armillaria mellea using synthetic and plant based substrates. J. Appl. Microbiol. 2020, in press. [Google Scholar] [CrossRef]
- Brimner, T.A.; Boland, G.J. A review of the non-target effects of fungi used to biologically control plant diseases. Agric. Ecosyst. Environ. 2003, 100, 3–16. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Chapman, B.; Xiao, G. Inoculation of stumps with Hypholoma fasciculare as a possible means to control Armillaria root disease. Can. J. Bot. 2000, 78, 129–134. [Google Scholar] [CrossRef]
- Dowson, C.G.; Rayner, A.D.; Boddy, L. Inoculation of mycelial cord-forming basidiomycetes into woodland soil and litter I. Initial establishment. New Phytol. 1988, 109, 335–341. [Google Scholar] [CrossRef]
- Dowson, C.G.; Rayner, A.D.; Boddy, L. Inoculation of mycelial cord-forming basidiomycetes into woodland soil and litter II. Resource capture and persistence. New Phytol. 1988, 109, 343–349. [Google Scholar] [CrossRef]
- Pozo, M.J.; Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 2007, 10, 393–398. [Google Scholar] [CrossRef]
- Gallou, A.; Mosquera, H.P.; Cranenbrouck, S.; Suárez, J.P.; Declerck, S. Mycorrhiza induced resistance in potato plantlets challenged by Phytophthora infestans. Physiol. Mol. Plant Pathol. 2011, 76, 20–26. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Nair, A.; Kolet, S.P.; Thulasiram, H.V.; Bhargava, S. Systemic jasmonic acid modulation in mycorrhizal tomato plants and its role in induced resistance against Alternaria alternata. Plant Biol. 2015, 17, 625–631. [Google Scholar] [CrossRef]
- Bruisson, S.; Maillot, P.; Schellenbaum, P.; Walter, B.; Gindro, K.; Deglène-Benbrahim, L. Arbuscular mycorrhizal symbiosis stimulates key genes of the phenylpropanoid biosynthesis and stilbenoid production in grapevine leaves in response to downy mildew and grey mould infection. Phytochemistry 2016, 131, 92–99. [Google Scholar] [CrossRef]
- Eghbaltalab, M.; Gay, G.; Bruchet, G. Antagonisme entre 15 espèces de Basidio-mycètes et 3 champignons pathogènes de racines d’arbres. Bull. Soc. Linn. Lyon. 1975, 44, 203–229. [Google Scholar]
- Nogales, A.; Nieto, A.C.; Morell, V.E.; Marfà, V.; Pinós, M.C. In vitro interaction studies between Glomus intraradices and Armillaria mellea in vines. Span. J. Agric. Res. 2010, 1, 62–68. [Google Scholar] [CrossRef]
- Riffle, J.W. Effect of two mycophagous nematodes on Armillaria mellea root rot of Pinus ponderosa seedlings. Plant Dis. Rep. 1973, 57, 355–357. [Google Scholar]
- Cayrol, J.C.; Dubos, B.; Guillaumin, J.J. Etude préliminaire in vitro de l’agressivité de quelque nematodes mycophages vis-àvis de Trichoderma viride Pers., T. polysporum (Link. ex. Pers.) Rifaï et Armillaria mellea (Vahl) Karst. Ann. Phytopathol. 1978, 10, 177–185. [Google Scholar]
- Mankau, R.; Mankau, S.K. The role of mycophagous nematodes in the soil. The relationships of Aphelenchus avenae to phytopathogenic soil fungi. In Soil Organisms; Doeksen, J., van der Drift, J., Eds.; North Holland: Amsterdam, The Netherlands, 1963; pp. 271–280. [Google Scholar]
- Nicholas, W.S. The Biology of Free-Living Nematodes; Clarendon Press: Oxford, UK, 1984. [Google Scholar]
- Lartey, R.T. Dynamics of soil flora and fauna in biological control of soil inhabiting plant pathogens. Plant Pathol. J. 2006, 5, 125–142. [Google Scholar] [CrossRef]
- Kulik, M.M. The potential for using cyanobacteria (blue-green algae) and algae in the biological control of plant pathogenic bacteria and fungi. Eur. J. Plant Pathol. 1995, 101, 585–599. [Google Scholar] [CrossRef]
- Piccardi, R.; Frosini, A.; Tredici, M.R.; Margheri, M.C. Bioactivity in free-living and symbiotic cyanobacteria of the genus Nostoc. J. Appl. Phycol. 2000, 12, 543–547. [Google Scholar] [CrossRef]
- Biondi, N.; Piccardi, R.; Margheri, M.C.; Rodolfi, L.; Smith, G.D.; Tredici, M.R. Evaluation of Nostoc strain ATCC 53789 as a potential source of natural pesticides. Appl. Environ. Microbiol. 2004, 70, 3313–3320. [Google Scholar] [CrossRef] [Green Version]
- Patterson, G.M.; Baldwin, C.L.; Bolis, C.M.; Caplan, F.R.; Karuso, H.; Larsen, L.K.; Levine, I.A.; Moore, R.E.; Nelson, C.S.; Tschappat, K.D.; et al. Antineoplastic activity of cultured blue-green algae (cyanophyta) 1. J. Phycol. 1991, 27, 530–536. [Google Scholar] [CrossRef]
- Panda, D.; Himes, R.H.; Moore, R.E.; Wilson, L.; Jordan, M.A. Mechanism of action of the unusually potent microtubule inhibitor cryptophycin 1. Biochemistry 1997, 36, 12948–12953. [Google Scholar] [CrossRef]
- Inderjit; van der Putten, W.H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 2010, 25, 512–519. [Google Scholar] [CrossRef]
- Belnap, J.; Phillips, S.L.; Sherrod, S.K.; Moldenke, A. Soil biota can change after exotic plant invasion: Does this affect ecosystem processes? Ecology 2005, 86, 3007–3017. [Google Scholar] [CrossRef]
- Mangla, S.; Callaway, R.M. Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 2008, 96, 58–67. [Google Scholar] [CrossRef]
- Anžlovar, S.; Koce, J.D. Antibacterial and antifungal activity of aqueous and organic extracts from indigenous and invasive species of goldenrod (Solidago spp.) grown in Slovenia. Phyton 2014, 54, 135–147. [Google Scholar] [CrossRef]
- Omirou, M.; Rousidou, C.; Bekris, F.; Papadopoulou, K.K.; Menkissoglou-Spiroudi, U.; Ehaliotis, C.; Karpouzas, D.G. The impact of biofumigation and chemical fumigation methods on the structure and function of the soil microbial community. Microb. Ecol. 2011, 61, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Baldi, E.; Toselli, M.; Malaguti, L.; Lazzeri, L. Evaluation of the biocidal effects of Brassica seed meal on Armillaria mellea. Ann. Appl. Biol. 2015, 167, 364–372. [Google Scholar] [CrossRef]
- Beal, E.J.; Henricot, B.; Peace, A.J.; Waghorn, I.A.G.; Denton, J.O. The action of allicin against Armillaria spp. in vitro. For. Pathol. 2015, 45, 450–458. [Google Scholar] [CrossRef]
- Reaves, J.L. Effects of Ash Leachates on Growth and Development of Armillaria Mellea in Culture; US Department of Agriculture, Forest Service, Pacific Northwest Forest and Range Experiment Station: Portland, OR, USA, 1984.
- Reaves, J.L. Interaction between Armillaria Root Disease, Trichoderma and Prescribed Fire in a Ponderosa Pine Forest. Ph.D. Thesis, Atlanta University, Atlanta, GA, USA, 1985. [Google Scholar]
- Gutter, W.D.; Baumgartner, K.; Browne, G.T.; Eskalen, A.; Latham, S.R.; Petit, E.; Bayramian, L.A. Root diseases of grapevines in California and their control. Australas. Plant Pathol. 2004, 33, 157–165. [Google Scholar] [CrossRef]
- Przybyl, K.; Manka, M. Influence of mineral salts upon activity of Trichoderma harzianum non-volatile metabolites on Armillaria spp. rhizomorphs. Acta Soc. Bot. Pol. 2004, 73, 327–330. [Google Scholar] [CrossRef] [Green Version]
- Ohr, H.D.; Munnecke, D.E. Effects of methyl bromide on antibiotic production by Armillaria mellea. Trans. Br. Mycol. Soc. 1974, 62, 65–72. [Google Scholar] [CrossRef]
- Raziq, F.; Fox, R.T. The integrated control of Armillaria mellea 1. Glasshouse experiments. Biol. Agric. Horticult. 2006, 23, 225–234. [Google Scholar] [CrossRef]
- Munnecke, D.E.; Kolbezen, M.J.; Wilbur, W.D. Effect of methyl bromide or carbon disulfide on Armillaria and Trichoderma growing on agar medium and relation to survival of Armillaria in soil following fumigation. Phytopathology 1973, 63, 1352–1357. [Google Scholar] [CrossRef]
- Munnecke, D.E.; Kolbezen, M.J.; Wilbur, W.D.; Ohr, H.D. Interactions involved in controlling Armillaria mellea. Plant Dis. 1981, 65, 384–389. [Google Scholar] [CrossRef]
- Ohr, H.D.; Munnecke, D.E.; Bricker, J.L. The interaction of Armillaria mellea and Trichoderma spp. as modified by methyl bromide. Phytopathology 1973, 63, 965–973. [Google Scholar] [CrossRef]
- Garrett, S.D. Effect of a soil microflora selected by carbon disulphide fumigation on survival of Armillaria mellea in woody host tissues. Can. J. Microbiol. 1957, 3, 135–149. [Google Scholar] [CrossRef]


| Co-occurring Insect Pest | Armillarioid Species | Location | Host Tree | Year | Reference |
|---|---|---|---|---|---|
| Lepidoptera | |||||
| Gypsy moth (Lymantria dispar) | Armillaria sp. | Massachusetts, USA | Oak | 1912–1921 | [205] |
| Armillaria sp. | New Jersey, USA | Oak | 1967 | [206] | |
| Armillaria sp. | Pennsylvania, USA | Oak | 1985 | [207] | |
| Armillaria sp. | Maryland, USA | Oak | 1985–1987 | [208] | |
| A. gallica | Pennsylvania, West Virginia, Maryland, USA | Oak | 1993 | [209] | |
| Eastern spruce budworm (Choristoneura fumiferana) | A. mellea sensu lato | Canada | Balsam fir | 1955–1958 | [210] |
| Armillaria spp. | New Brunswick, Canada | Balsam fir | 1960s | [211] | |
| Armillaria spp. | Newfoundland, Canada | Black spruce | early 1980s | [212] | |
| Western spruce budworm (Choristoneura occidentalis) | A. altimontana (NABS X) | Eastern Oregon, USA | Grand fir | 1989 | [213] |
| Maple webworm (Tetralopha asperatella) | A. mellea sensu lato | Wisconsin, USA | Sugar maple | late 1950s | [214] |
| Oak leaf tier (Acleris semipurpurana) | Armillaria sp. | Pennsylvania, USA | Red oak Scarlet oak | 1960s | [215] |
| Saddled prominent caterpillar (Heterocampa guttavitta) | Armillaria spp. | North-Central New York, USA | Sugar maple | 1991 | [194] |
| European spruce needleminer (Epinotia nanaxa) | Armillaria sp. | Norway | Norway spruce | 1984 | [216] |
| Linden loopers (Erranis tiliaria) | Armillaria sp. | Maryland, USA | Oak | 1985–1987 | [208] |
| Coleoptera | |||||
| Eight-toothed European spruce bark beetle | A. mellea sensu lato | South Poland | Spruce | 1974 | [217] |
| (Ips typographus) | Armillaria sp. | Northeastern Slovakia | Spruce | 1995 | [218] |
| Armillaria spp. | Western Beskidy, Poland | Norway spruce | 2000s | [219] | |
| A. ostoyae A. cepistipes A. borealis | Bohemian forest, Czech Republic | Norway spruce | 2002 | [220] | |
| A. ostoyae A. cepistipes A. gallica | Eastern Czech Republic | Norway spruce | 2010s | [183] | |
| Fir engraver (Scolytus ventralis) | A. mellea sensu lato | Northern Idaho, USA | Grand fir | 1972 | [221] |
| Armillaria sp. | Idaho, USA | Grand fir | 1974 | [222] | |
| A. mellea sensu lato | Colorado, USA | White fir | 1981 | [223] | |
| A. mellea sensu lato | Oregon and Washington, USA | Grand fir Rocky Mountain white fir Pacific silver fir Noble fir California red fir Subalpine fir | 1977 | [224] | |
| Western balsam bark beetle (Dryocoetes confusus) | A. mellea sensu lato | Oregon and Washington, USA | Subalpine fir | 1977 | [224] |
| A. solidipes | Central Oregon, USA | Grand fir | 1979, 1992 | [225] | |
| A. mellea sensu lato | Colorado, USA | Subalpine fir | 1981 | [223] | |
| Warren’s rootcollar weevil (Hylobius warreni) | A. mellea sensu lato | Saskatchewan, Canada | White spruce | 1960 | [226] |
| Armillaria sp. | Newfoundland, Canada | Sitka spruce Norway spruce Red pine | 1970 | [227] | |
| Mountain pine beetle (Dendroctonus ponderosae) | A. mellea sensu lato | Idaho, USA | Western white pine | 1975 | [228] |
| A. mellea sensu lato | Utah, USA | Lodgepole pine | 1983 | [229] | |
| Bark beetle (Pityogenes fossifrons) | A. mellea sensu lato | Idaho, USA | Western white pine | 1975 | [230] |
| Double-spined bark beetle (Ips duplicatus) Sixtoothed spruce bark beetle (Pityogenes chalcographus) | A. ostoyae A. cepistipes A. gallica | Eastern Czech Republic | Norway spruce | 2010s | [183] |
| Twolined chestnut borer (Agrilus bilineatus) | A. mellea sensu lato | Connecticut, USA | Oak | 1969–1973 | [231] |
| Spruce wood engraver (Pityogenes chalcographus) Brown longhorn beetle (Obrium brunneum) Spruce shortwing beetle (Molorchus minor) Pogonocherus fasciculatus Phthorophloeus spinulosus | Armillaria sp. | Western Carpathian mountain, Czech Republic | Norway spruce | 1990s | [232] |
| Hemiptera | |||||
| Balsam woolly adelgid (Adelges piceae) | A. mellea sensu lato | Newfoundland, Canada | Balsam fir | 1960s | [233,234] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kedves, O.; Shahab, D.; Champramary, S.; Chen, L.; Indic, B.; Bóka, B.; Nagy, V.D.; Vágvölgyi, C.; Kredics, L.; Sipos, G. Epidemiology, Biotic Interactions and Biological Control of Armillarioids in the Northern Hemisphere. Pathogens 2021, 10, 76. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10010076
Kedves O, Shahab D, Champramary S, Chen L, Indic B, Bóka B, Nagy VD, Vágvölgyi C, Kredics L, Sipos G. Epidemiology, Biotic Interactions and Biological Control of Armillarioids in the Northern Hemisphere. Pathogens. 2021; 10(1):76. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10010076
Chicago/Turabian StyleKedves, Orsolya, Danish Shahab, Simang Champramary, Liqiong Chen, Boris Indic, Bettina Bóka, Viktor Dávid Nagy, Csaba Vágvölgyi, László Kredics, and György Sipos. 2021. "Epidemiology, Biotic Interactions and Biological Control of Armillarioids in the Northern Hemisphere" Pathogens 10, no. 1: 76. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens10010076