Chemoreversal Agents from Taiwanofungus Genus and Their More Potent Methyl Derivatives Targeting Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Cytotoxic Evaluation of Compounds 5–28 on Hela S3 and KBvin Cells
2.3. Collateral Sensitivity Evaluation of the Effects of Zhankuic Acid Type Compounds and Their Methyl Esters on Paclitaxel Cytotoxicity
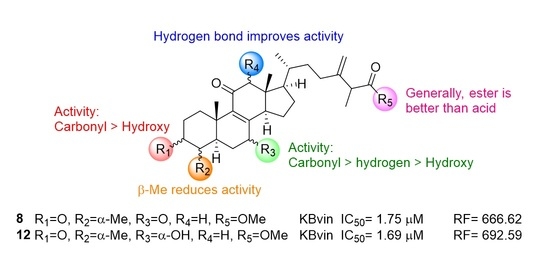
2.4. Research on the Mechanism of the Most Active Compounds
3. Materials and Methods
3.1. General
3.2. General Procedure for Methylation
3.3. Methyl 3α,7α,12α-Trihydroxy-4α-methylergosta-8,24(28)-dien-11-on-26-oate (6)
3.4. Zhankuic Acid A Methyl Ester (8)
3.5. Methyl 7α, 12α-Dihydroxy-3,11-dioxo-4α-methylergosta-8,24(28)-dien-26-oate (10)
3.6. Methyl-7β-hydroxy-3,11-dioxo-4α-methylergosta-8,24(28)-dien-26-oate (12)
3.7. Methyl 3α,4β,7β-Trihydroxyergosta-8,24(28)-dien-11-on-26-oate (methyl antcamphorol D, 14)
3.8. Methyl 3α,12α-Dihydroxy 4α-methylergosta-8,24(28)-diene-7,11-dion-26-oate (methyl antcinate H, 16)
3.9. Zhankuic Acid Methyl Ester C 3-O-formate (18)
3.10. Methyl 3α-Hydroxy-7,11-dioxo-4α-methylergosta-8,24(28)-dien-26-oate (20)
3.11. Methyl 7β-Hydroxy-3,11-dioxo-4α-methylergosta-8,24(28)-dien-26-oate (22)
3.12. Methyl 4α-Methylergosta-8,24(28)-diene-3,11-dion-26-oate (24)
3.13. Methyl 3α,7β-Dihydroxy-4α-methylergosta-8,24(28)-dien-11-on-26-oate (26)
3.14. Methyl 3β,12β-Dihydroxy-11-oxo-4β-methylergosta-8,24(28)-dien-26-oate (methyl antcin M, 28)
3.15. Culture of Cell Lines
3.16. SRB Cytotoxicity Assay and Reversal Fold Calculation
3.17. Enzyme-Linked Immunosorbent Assay (ELISA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gottesman, M.M. Mechanisms of Cancer Drug Resistance. Annu. Rev. Med. 2002, 53, 615–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holohan, C.; Schaeybroeck, S.V.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; Nie, Y.; Shi, Y.; Fan, D. Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett. 2014, 347, 159–166. [Google Scholar] [CrossRef]
- Chabner, B.A.; Roberts, T.G.J. Timeline: Chemotherapy and the war on cancer. Nat. Rev. Cancer 2005, 5, 65–72. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Luqmani, Y.A. Mechanisms of Drug Resistance in Cancer Chemotherapy. Med. Princ. Pract. 2005, 14, 35–48. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amawi, H.; Sim, H.M.; Tiwari, A.K.; Ambudkar, S.V.; Shukla, S. ABC Transporter-Mediated Multidrug-Resistant Cancer. Adv. Exp. Med. Biol. 2019, 1141, 549–580. [Google Scholar] [CrossRef]
- Callaghan, R.; Luk, F.; Bebawy, M. Inhibition of the multidrug resistance P-glycoprotein: Time for a change of strategy? Drug Metab. Dispos. 2014, 42, 623–631. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Leite de Oliveira, R.; Huijberts, S.; Bosdriesz, E.; Pencheva, N.; Brunen, D.; Bosma, A.; Song, J.Y.; Zevenhoven, J.; Los-de Vries, G.T.; et al. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell 2018, 173, 1413–1425 e14. [Google Scholar] [CrossRef] [Green Version]
- Efferth, T.; Saeed, M.E.M.; Kadioglu, O.; Seo, E.J.; Shirooie, S.; Mbaveng, A.T.; Nabavi, S.M.; Kuete, V. Collateral sensitivity of natural products in drug-resistant cancer cells. Biotechnol. Adv. 2020, 38, 107342. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Teng, Y.-N.; Lin, K.-I.; Wang, C.C.N.; Morris-Natschke, S.L.; Lee, K.-H.; Hung, C.-C. Danazol mediates collateral sensitivity via STAT3/Myc related pathway in multidrug-resistant cancer cells. Sci. Rep. 2019, 9, 11628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barré, B.; Vigneron, A.; Perkins, N.; Roninson, I.B.; Gamelin, E.; Coqueret, O. The STAT3 oncogene as a predictive marker of drug resistance. Trends Mol. Med. 2007, 13, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. The JAK-STAT signaling pathway: Input and output integration. J. Immunol. 2007, 178, 2623–2629. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.-Q.; Liu, B.; Wang, Y.-P.; Li, J.-K.; Zhu, P.-L.; Li, T.; Tse, K.-W.; Chou, J.-Y.; Yin, C.-L.; Bai, J.-X.; et al. Activation of STAT3 is a key event in TLR4 signaling-mediated melanoma progression. Cell Death Dis. 2020, 11, 246. [Google Scholar] [CrossRef]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, J.; Wu, J.; Zhong, S.; Li, H. Inhibition of JAK2 Reverses Paclitaxel Resistance in Human Ovarian Cancer Cells. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2015, 25, 1557–1564. [Google Scholar] [CrossRef]
- Su, W.P.; Cheng, F.Y.; Shieh, D.B.; Yeh, C.S.; Su, W.C. PLGA nanoparticles codeliver paclitaxel and Stat3 siRNA to overcome cellular resistance in lung cancer cells. Int. J. Nanomed. 2012, 7, 4269–4283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Cao, J.; Wu, J.; Sullivan, K.; Shen, J.; Ryu, B.; Xu, Z.; Wei, W.; Cui, R. Stat3-Targeted Therapies Overcome the Acquired Resistance to Vemurafenib in Melanomas. J. Investig. Dermatol. 2013, 133, 2041–2049. [Google Scholar] [CrossRef] [Green Version]
- Girotti, M.R.; Pedersen, M.; Sanchez-Laorden, B.; Viros, A.; Turajlic, S.; Niculescu-Duvaz, D.; Zambon, A.; Sinclair, J.; Hayes, A.; Gore, M.; et al. Inhibiting EGF Receptor or SRC Family Kinase Signaling Overcomes BRAF Inhibitor Resistance in Melanoma. Cancer Discov. 2013, 3, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Lai, I.C.; Lai, G.M.; Chow, J.M.; Lee, H.L.; Yeh, C.F.; Li, C.H.; Yan, J.L.; Chuang, S.E.; Whang-Peng, J.; Bai, K.J.; et al. Active fraction (HS7) from Taiwanofungus camphoratus inhibits AKT-mTOR, ERK and STAT3 pathways and induces CDK inhibitors in CL1-0 human lung cancer cells. Chin. Med. 2017, 12, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.N.; Wang, Y.H.; Wu, T.S.; Hung, H.Y.; Hung, C.C. Zhankuic Acids A, B and C from Taiwanofungus Camphoratus Act as Cytotoxicity Enhancers by Regulating P-Glycoprotein in Multi-Drug Resistant Cancer Cells. Biomolecules 2019, 9, 759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.-J.; Leu, Y.-L.; Chen, C.-H.; Chao, C.-H.; Shen, D.-Y.; Chan, H.-H.; Lee, E.J.; Wu, T.-S.; Wang, Y.-H.; Shen, Y.-C.; et al. Camphoratins A−J, Potent Cytotoxic and Anti-inflammatory Triterpenoids from the Fruiting Body of Taiwanofungus camphoratus. J. Nat. Prod. 2010, 73, 1756–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.-S.; Chao, C.-H.; Shen, D.-Y.; Chan, H.-H.; Chen, C.-H.; Liao, Y.-R.; Wu, S.-J.; Leu, Y.-L.; Shen, Y.-C.; Kuo, Y.-H.; et al. Biologically active constituents from the fruiting body of Taiwanofungus camphoratus. Bioorganic Med. Chem. 2011, 19, 677–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.-C.; Chen, C.-F.; Hung, C.-C.; Lam, S.-H.; Hung, H.-Y.; Li, Y.-C.; Chen, F.-A.; Shieh, P.-C.; Kuo, P.-C.; Wu, T.-S. Bioactive naphthoquinones and triterpenoids from the fruiting bodies of Taiwanofungus salmoneus. Bioorganic Chem. 2021, 112, 104939. [Google Scholar] [CrossRef]
- Chen, C.-H.; Yang, S.-W.; Shen, Y.-C. New Steroid Acids from Antrodia cinnamomea, a Fungal Parasite of Cinnamomum micranthum. J. Nat. Prod. 1995, 58, 1655–1661. [Google Scholar] [CrossRef]
- Hung, H.Y.; Hung, C.C.; Liang, J.W.; Chen, C.F.; Chen, H.Y.; Shieh, P.C.; Kuo, P.C.; Wu, T.S. Constituents and Anti-Multidrug Resistance Activity of Taiwanofungus camphoratus on Human Cervical Cancer Cells. Molecules 2019, 24, 3730. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.F.; Wang, S.H.; Chang, S.J.; Shiau, A.L.; Her, L.S.; Shieh, G.S.; Chen, C.F.; Chang, C.C.; Su, Y.C.; Wu, C.L.; et al. Zhankuic acid A as a novel JAK2 inhibitor for the treatment of concanavalin A-induced hepatitis. Biochem. Pharmacol. 2014, 91, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Wang, Q.; Ji, S.; Huang, Y.; Liu, K.D.; Zhang, Z.X.; Bo, T.; Tzeng, Y.M.; Guo, D.A.; Ye, M. Metabolites identification and multi-component pharmacokinetics of ergostane and lanostane triterpenoids in the anticancer mushroom Antrodia cinnamomea. J. Pharm. Biomed. Anal. 2015, 111, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Kuang, Y.; He, J.-B.; Tang, R.; Xu, L.-L.; Leung, C.-H.; Ma, D.-L.; Qiao, X.; Ye, M. Antcamphorols A–K, Cytotoxic and ROS Scavenging Triterpenoids from Antrodia camphorata. J. Nat. Prod. 2020, 83, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-C.; Wang, Y.-H.; Chang, T.-T.; Lin, L.-C.; Don, M.-J.; Hou, Y.-C.; Liou, K.-T.; Chang, S.; Wang, W.-Y.; Ko, H.-C.; et al. Anti-Inflammatory Ergostanes from the Basidiomata of Antrodia salmonea. Planta Med. 2007, 73, 1208–1213. [Google Scholar] [CrossRef] [PubMed]









| No | Name | Reference |
|---|---|---|
| 5 | Salmonone D | [27] |
| 7 | Antcin B | [32] |
| 9 | Salmonone E | [27] |
| 11 | 7α-hydroxy-3,11-dioxo-4α-methylergosta-8,24(28)-dien-26-oic acid | [25] |
| 13 | Antcin K | [33] |
| 15 | Zhankuic acid C | [25] |
| 17 | Zhankuic acid C 3-O-formate | [27] |
| 19 | Zhankuic acid B | [32] |
| 21 | Antcin C | [32] |
| 23 | Antcin A | [25] |
| 25 | Camphoratin B | [25] |
| 27 | 3α,12α-dihydroxy-4α-methylergosta-8,24(28)-dien-11-on-26-oic acid | [33] |

| No | Structure | KBvin | HeLa S3 | ||||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R5 | IC50 (μM) | IC50 (μM) | |
| 5 | α-OH | α-Me | α-OH | α-OH | OH | >40 | >40 |
| 6 | α-OH | α-Me | α-OH | α-OH | OMe | 19.83 ± 0.33 | 21.78 ± 1.13 |
| 7 | =O | α-Me | =O | H | OH | 32.88 ± 1.16 | >40 |
| 8 | =O | α-Me | =O | H | OMe | 24.44 ± 0.34 | 27.14 ± 1.36 |
| 9 | =O | α-Me | α-OH | α-OH | OH | >40 | >40 |
| 10 | =O | α-Me | α-OH | α-OH | OMe | 21.77 ± 0.86 | >40 |
| 11 | =O | α-Me | α-OH | H | OH | 32.41 ± 1.27 | >40 |
| 12 | =O | α-Me | α-OH | H | OMe | 22.29 ± 0.19 | 29.00 ± 0.55 |
| 13 | α-OH | α-Me, β-OH | β-OH | H | OH | >40 | >40 |
| 14 | α-OH | α-Me, β-OH | β-OH | H | OMe | >40 | >40 |
| 15 | α-OH | α-Me | =O | α-OH | OH | >40 | >40 |
| 16 | α-OH | α-Me | =O | α-OH | OMe | 28.23 ± 0.01 | 27.26 ± 1.66 |
| 17 | α-OCOH | α-Me | =O | α-OH | OH | >40 | >40 |
| 18 | α-OCOH | α-Me | =O | α-OH | OMe | 20.95 ± 3.54 | >40 |
| 19 | α-OH | α-Me | =O | H | OH | >40 | >40 |
| 20 | α-OH | α-Me | =O | H | OMe | >40 | >40 |
| 21 | =O | α-Me | β-Me | H | OH | >40 | >40 |
| 22 | =O | α-Me | β-Me | H | OMe | 14.48 ± 1.00 | 26.79 ± 0.99 |
| 23 | =O | α-Me | H | H | OH | >40 | >40 |
| 24 | =O | α-Me | H | H | OMe | >40 | >40 |
| 25 | α-OH | α-Me | β-OH | H | OH | >40 | >40 |
| 26 | α-OH | α-Me | β-OH | H | OMe | 27.32 ± 0.33 | >40 |
| 27 | α-OH | α-Me | H | α-OH | OH | >40 | >40 |
| 28 | α-OH | α-Me | H | α-OH | OMe | 30.98 ± 2.47 | 31.84 ± 2.67 |
| Paclitaxel | 1168.50 ± 75.02 | 12.11 ± 2.04 | |||||

| No | Structure | Conc. (μM) | KBvin | HeLa S3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | RF 1 | IC50 (nM) | RF 1 | |||||||
| Paclitaxel only | - | 1168.50 ± 75.02 | 1.00 | 12.11 ± 2.04 | 1.00 | |||||
| R1 | R2 | R3 | R4 | R5 | - | |||||
| 5 | α-OH | α-Me | α-OH | α-OH | OH | 10 | 1078.64 ± 77.01 | 1.08 | 7.96 ± 0.21 * | 1.52 |
| 20 | 972.99 ± 37.93 * | 1.20 | 7.53 ± 0.47 * | 1.61 | ||||||
| 6 | α-OH | α-Me | α-OH | α-OH | OMe | 5 | 89.06 ± 0.43 * | 13.12 | 4.76 ± 0.01 * | 2.55 |
| 10 | 18.25 ± 4.97 * | 64.02 | 4.13 ± 0.60 * | 4.49 | ||||||
| 7 | =O | α-Me | =O | H | OH | 10 | 613.33 ± 2.78 * | 1.91 | 7.94 ± 0.63 * | 1.53 |
| 20 | 249.51 ± 23.44 * | 4.68 | 6.67 ± 0.27 * | 1.82 | ||||||
| 8 | =O | α-Me | =O | H | OMe | 5 | 58.41 ± 6.46 * | 20.01 | 4.73 ± 0.15 * | 2.56 |
| 10 | 1.75 ± 0.63 * | 666.62 | 2.70 ± 1.32 * | 4.49 | ||||||
| 9 | =O | α-Me | α-OH | α-OH | OH | 10 | 1075.33 ± 83.81 | 1.09 | 7.55 ± 0.63* | 1.60 |
| 20 | 974.82 ± 60.89 | 1.20 | 8.03 ± 0.12* | 1.51 | ||||||
| 10 | =O | α-Me | α-OH | α-OH | OMe | 5 | 77.86 ± 7.25 * | 15.01 | 4.43 ± 0.26 * | 2.73 |
| 10 | 52.92 ± 7.90 * | 22.08 | 3.08 ± 1.20 * | 3.93 | ||||||
| 11 | =O | α-Me | α-OH | H | OH | 10 | 730.30 ± 29.35 * | 1.60 | 8.33 ± 0.16 * | 1.45 |
| 20 | 279.61 ± 7.84 * | 4.18 | 6.51 ± 0.25 * | 1.86 | ||||||
| 12 | =O | α-Me | α-OH | H | OMe | 5 | 171.88 ± 73.04 * | 6.80 | 4.65 ± 0.08 * | 2.60 |
| 10 | 1.69 ± 0.17 * | 692.59 | 2.61 ± 1.37 * | 4.64 | ||||||
| 13 | α-OH | α-Me, β-OH | β-OH | H | OH | 10 | 987.07 ± 75.18 | 1.18 | 17.93 ± 2.86 | 0.68 |
| 20 | 902.61 ± 7.57 * | 1.29 | 9.11 ± 0.16 | 1.33 | ||||||
| 14 | α-OH | α-Me, β-OH | β-OH | H | OMe | 5 | 664.92 ± 38.20 * | 1.76 | 5.09 ± 0.19 * | 2.38 |
| 10 | 293.99 ± 57.13 * | 3.97 | 4.23 ± 1.42 * | 2.86 | ||||||
| 15 | α-OH | α-Me | =O | α-OH | OH | 10 | 931.63 ± 24.81 * | 1.25 | 15.44 ± 1.37 | 0.78 |
| 20 | 722.92 ± 23.34 * | 1.62 | 9.49 ± 0.62 | 1.28 | ||||||
| 16 | α-OH | α-Me | =O | α-OH | OMe | 5 | 63.35 ± 6.10 * | 18.44 | 5.00 ± 0.06 * | 2.42 |
| 10 | 4.94 ± 3.31 * | 236.38 | 3.36 ± 0.42 * | 3.60 | ||||||
| 17 | α-OCOH | α-Me | =O | α-OH | OH | 10 | 729.20 ± 13.82 * | 1.60 | 5.07 ± 0.75 * | 2.39 |
| 20 | 659.03 ± 28.57 * | 1.77 | 4.99 ± 1.39 * | 2.43 | ||||||
| 18 | α-OCOH | α-Me | =O | α-OH | OMe | 5 | 41.12 ± 9.34 * | 28.41 | 4.01 ± 0.01 * | 3.02 |
| 10 | 26.08 ± 7.86 * | 44.81 | 2.33 ± 0.78 * | 5.21 | ||||||
| 19 | α-OH | α-Me | =O | H | OH | 10 | 682.65 ± 32.57 * | 1.71 | 4.29 ±1.08 * | 2.82 |
| 20 | 564.84 ± 25.45 * | 2.07 | 4.19 ± 1.40 * | 2.89 | ||||||
| 20 | α-OH | α-Me | =O | H | OMe | 5 | 444.16 ± 44.85 * | 2.63 | 4.89 ± 0.18 * | 2.47 |
| 10 | 103.47 ± 7.32 * | 11.29 | 3.47 ± 0.32 * | 3.49 | ||||||
| 21 | =O | α-Me | β-Me | H | OH | 10 | 769.34 ± 8.16 * | 1.52 | 4.86 ± 0.51 * | 2.49 |
| 20 | 657.20 ± 30.86 * | 1.78 | 4.60 ± 1.44 * | 2.63 | ||||||
| 22 | =O | α-Me | β-Me | H | OMe | 5 | 17.80 ± 6.56 * | 65.66 | 2.94 ± 0.22 * | 4.12 |
| 10 | 108.96 ± 15.25 * | 10.72 | 0.51 ± 0.10 * | 23.64 | ||||||
| 23 | =O | α-Me | H | H | OH | 10 | 669.22 ± 39.57 * | 1.75 | 4.65 ± 0.55 * | 2.60 |
| 20 | 461.90 ± 46.61 * | 2.53 | 4.26 ± 1.08 * | 2.85 | ||||||
| 24 | =O | α-Me | H | H | OMe | 5 | 544.76 ± 24.01 * | 2.14 | 4.62 ± 0.02 * | 2.62 |
| 10 | 136.03 ± 27.36 * | 8.59 | 4.59 ± 0.48 * | 2.64 | ||||||
| 25 | α-OH | α-Me | β-OH | H | OH | 10 | 852.31 ± 47.93 * | 1.37 | 5.10 ± 0.36 * | 2.37 |
| 20 | 723.07 ± 6.95 * | 1.62 | 5.07 ± 0.31 * | 2.39 | ||||||
| 26 | α-OH | α-Me | β-OH | H | OMe | 5 | 6.33 ± 0.10 * | 184.47 | 3.55 ± 0.29 * | 3.41 |
| 10 | 3.35 ± 1.00 * | 348.80 | 3.38 ± 0.25 * | 3.58 | ||||||
| 27 | α-OH | α-Me | H | α-OH | OH | 10 | 681.06 ± 12.91 * | 1.72 | 5.42 ± 0.01 * | 2.23 |
| 20 | 558.44 ± 4.62 * | 2.09 | 5.33 ± 0.04 * | 2.27 | ||||||
| 28 | α-OH | α-Me | H | α-OH | OMe | 5 | 65.90 ± 5.37 * | 17.73 | 4.68 ± 0.01 * | 2.59 |
| 10 | 3.93 ± 1.96 * | 297.23 | 4.02 ± 0.01 * | 3.01 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, K.-H.; Hung, C.-C.; Wu, T.-S.; Chen, C.-F.; Wu, I.-T.; Kuo, P.-C.; Lam, S.-H.; Hung, H.-Y. Chemoreversal Agents from Taiwanofungus Genus and Their More Potent Methyl Derivatives Targeting Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation. Pharmaceuticals 2021, 14, 916. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090916
Yu K-H, Hung C-C, Wu T-S, Chen C-F, Wu I-T, Kuo P-C, Lam S-H, Hung H-Y. Chemoreversal Agents from Taiwanofungus Genus and Their More Potent Methyl Derivatives Targeting Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation. Pharmaceuticals. 2021; 14(9):916. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090916
Chicago/Turabian StyleYu, Ko-Hua, Chin-Chuan Hung, Tian-Shung Wu, Chin-Fu Chen, I-Ting Wu, Ping-Chung Kuo, Sio-Hong Lam, and Hsin-Yi Hung. 2021. "Chemoreversal Agents from Taiwanofungus Genus and Their More Potent Methyl Derivatives Targeting Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation" Pharmaceuticals 14, no. 9: 916. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090916