Regulation of m6A Methylation as a New Therapeutic Option against COVID-19
Abstract
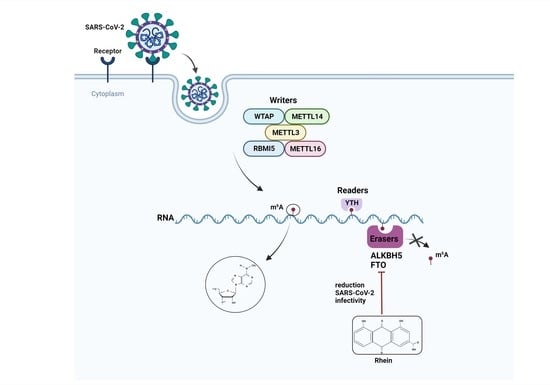
:1. Introduction
2. Results
2.1. Cytotoxicity Assay
2.2. Antiviral Activity
2.3. RNA m6A Quantification
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Viruses
4.3. Cytotoxicity Assay
4.4. Antiviral Assay: Co-Exposure
4.5. m6A Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohan, B.; Nambiar, V. COVID-19: An insight into SARS-CoV-2 pandemic originated at Wuhan City in Hubei Province of China. J. Infect. Dis. Epidemiol. 2020, 6, 146. [Google Scholar] [CrossRef]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard; World Health Organization: Roma, Italy, 2021. [Google Scholar]
- Iacob, S.; Iacob, D.G. SARS-CoV-2 Treatment Approaches: Numerous Options, No Certainty for a Versatile Virus. Front. Pharmacol. 2020, 11, 1224. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.W.; Xie, Y.; Tang, L.S.; Pu, D.; Zhu, Y.J.; Liu, J.Y.; Ma, X.L. Therapeutic targets and interventional strategies in COVID-19: Mechanisms and clinical studies. Signal Transduct. Target. Ther. 2021, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Trindade, G.G.; Caxito, S.M.C.; Xavier, A.; Xavier, M.A.S.; BrandAo, F. COVID-19: Therapeutic approaches description and discussion. An. Acad. Bras. Cienc. 2020, 92, e20200466. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, M.; Galdiero, M.; Folliero, V.; Zannella, C.; De Filippis, A.; Mali, A.; Rinaldi, L.; Franci, G. SARS-CoV-2 vaccine development: Where are we? Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 2752–2784. [Google Scholar] [CrossRef]
- Chakraborty, S.; Mallajosyula, V.; Tato, C.M.; Tan, G.S.; Wang, T.T. SARS-CoV-2 vaccines in advanced clinical trials: Where do we stand? Adv. Drug. Deliv. Rev. 2021, 172, 314–338. [Google Scholar] [CrossRef]
- Li, S.; Kong, L.; Yu, X.; Zheng, Y. Host-virus interactions: From the perspectives of epigenetics. Rev. Med. Virol. 2014, 24, 223–241. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [Green Version]
- Milavetz, B.I.; Balakrishnan, L. Viral epigenetics. Methods Mol. Biol. 2015, 1238, 569–596. [Google Scholar] [CrossRef]
- Nehme, Z.; Pasquereau, S.; Herbein, G. Control of viral infections by epigenetic-targeted therapy. Clin. Epigenetics 2019, 11, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, K.; Cullen, B.R. Epigenetic and epitranscriptomic regulation of viral replication. Nat. Rev. Microbiol. 2020, 18, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenetics 2020, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef]
- Beacon, T.H.; Su, R.C.; Lakowski, T.M.; Delcuve, G.P.; Davie, J.R. SARS-CoV-2 multifaceted interaction with the human host. Part II: Innate immunity response, immunopathology, and epigenetics. IUBMB Life 2020, 72, 2331–2354. [Google Scholar] [CrossRef]
- Kianmehr, A.; Faraoni, I.; Kucuk, O.; Mahrooz, A. Epigenetic alterations and genetic variations of angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV-2: Potential clinical implications. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1587–1598. [Google Scholar] [CrossRef]
- Corley, M.J.; Ndhlovu, L.C. DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Preprints 2020, 2020, 030295. [Google Scholar]
- Asada, K.; Bolatkan, A.; Takasawa, K.; Komatsu, M.; Kaneko, S.; Hamamoto, R. Critical Roles of N(6)-Methyladenosine (m(6)A) in Cancer and Virus Infection. Biomolecules 2020, 10, 1071. [Google Scholar] [CrossRef]
- Williams, G.D.; Gokhale, N.S.; Horner, S.M. Regulation of Viral Infection by the RNA Modification N6-Methyladenosine. Annu. Rev. Virol. 2019, 6, 235–253. [Google Scholar] [CrossRef]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m(6)A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef]
- Zhang, X.; Hao, H.; Ma, L.; Zhang, Y.; Hu, X.; Chen, Z.; Liu, D.; Yuan, J.; Hu, Z.; Guan, W. Methyltransferase-like 3 Modulates Severe Acute Respiratory Syndrome Coronavirus-2 RNA N6-Methyladenosine Modification and Replication. mBio 2021, 12, e0106721. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.P.; Li, K.; Ye, Q.; Zhou, H.Y.; Sun, H.; Li, X.; Yu, L.; Deng, Y.Q.; Li, R.T.; et al. The m(6)A methylome of SARS-CoV-2 in host cells. Cell Res. 2021, 31, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.M.; Depledge, D.P.; Thompson, L.; Srinivas, K.P.; Grande, R.C.; Vink, E.I.; Abebe, J.S.; Blackaby, W.P.; Hendrick, A.; Albertella, M.R.; et al. Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes. Dev. 2021, 35, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Zhang, Q.; Wang, K.; Zhang, X.; Yang, R.; Bi, K.; Chen, W.; Diao, H. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis. 2021, 12, 732. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Luo, G.; Chen, D.; Xiang, Z. A Comprehensive and System Review for the Pharmacological Mechanism of Action of Rhein, an Active Anthraquinone Ingredient. Front. Pharm. 2016, 7, 247. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.X.; Xia, W.; Yue, W.; Peng, C.; Rahman, K.; Zhang, H. Rhein: A Review of Pharmacological Activities. Evid. Based Complement. Altern. Med. 2015, 2015, 578107. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Ye, F.; Yu, L.; Jia, G.; Huang, X.; Zhang, X.; Peng, S.; Chen, K.; Wang, M.; Gong, S.; et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 2012, 134, 17963–17971. [Google Scholar] [CrossRef]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.; Song, H.; Zhu, J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018, 46, 1412–1423. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wu, Y.; Chen, C.; Gu, Y.; Zhu, C.; Wang, S.; Chen, J.; Zhang, L.; Lv, L.; Zhang, G.; et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta. Pharm. Sin. B 2021, 11, 222–236. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef] [Green Version]
- Brocard, M.; Ruggieri, A.; Locker, N. m6A RNA methylation, a new hallmark in virus-host interactions. J. Gen. Virol. 2017, 98, 2207–2214. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Hayakawa, A.; Sano, R.; Fukuda, H.; Harada, M.; Kubo, R.; Okawa, T.; Kominato, Y. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci. Rep. 2021, 11, 3379. [Google Scholar] [CrossRef]
- Beacon, T.H.; Delcuve, G.P.; Davie, J.R. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus(1). Genome 2021, 64, 386–399. [Google Scholar] [CrossRef]
- Ragia, G.; Manolopoulos, V.G. Assessing COVID-19 susceptibility through analysis of the genetic and epigenetic diversity of ACE2-mediated SARS-CoV-2 entry. Pharmacogenomics 2020, 21, 1311–1329. [Google Scholar] [CrossRef]
- Pitt, B.; Sutton, N.R.; Wang, Z.; Goonewardena, S.N.; Holinstat, M. Potential repurposing of the HDAC inhibitor valproic acid for patients with COVID-19. Eur. J. Pharmacol. 2021, 898, 173988. [Google Scholar] [CrossRef]
- Liu, K.; Zou, R.; Cui, W.; Li, M.; Wang, X.; Dong, J.; Li, H.; Li, H.; Wang, P.; Shao, X. Clinical HDAC inhibitors are effective drugs to prevent the entry of SARS-CoV2. ACS Pharmacol. Transl. Sci. 2020, 3, 1361–1370. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018, 172, 90–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Liu, J.; Xiao, W.; Zeng, Q.; Bo, H.; Zhu, Y.; Gong, L.; He, D.; Xing, X.; Li, R.; et al. SIRT1 Regulates N(6)-Methyladenosine RNA Modification in Hepatocarcinogenesis by Inducing RANBP2-Dependent FTO SUMOylation. Hepatology 2020, 72, 2029–2050. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Kandpal, M.; Zhao, G.; Cardenas, H.; Ji, Y.; Chaparala, A.; Tanner, E.J.; Chen, J.; Davuluri, R.V.; et al. FTO-Dependent N (6)-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 2020, 80, 3200–3214. [Google Scholar] [CrossRef]
- Song, H.; Feng, X.; Zhang, H.; Luo, Y.; Huang, J.; Lin, M.; Jin, J.; Ding, X.; Wu, S.; Huang, H.; et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019, 15, 1419–1437. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.C.; Yang, Q. NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 2020, 16, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Su, R.; Stanford, S.; Chen, J. Critical Enzymatic Functions of FTO in Obesity and Cancer. Front. Endocrinol. 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.W.; Su, Y.; Sheng, J.T.; Gu, L.M.; Zhao, Y.; Chen, X.X.; Chen, C.; Li, W.Z.; Li, K.S.; Dai, J.P. Anti-influenza A virus activity of rhein through regulating oxidative stress, TLR4, Akt, MAPK, and NF-kappaB signal pathways. PLoS ONE 2018, 13, e0191793. [Google Scholar] [CrossRef] [Green Version]
- Bracci, N.; Pan, H.C.; Lehman, C.; Kehn-Hall, K.; Lin, S.C. Improved plaque assay for human coronaviruses 229E and OC43. PeerJ 2020, 8, e10639. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Giugliano, R.; Chianese, A.; Buonocore, C.; Vitale, G.A.; Sanna, G.; Sarno, F.; Manzin, A.; Nebbioso, A.; Termolino, P. Antiviral Activity of Vitis vinifera Leaf Extract against SARS-CoV-2 and HSV-1. Viruses 2021, 13, 1263. [Google Scholar] [CrossRef] [PubMed]
- Squillaci, G.; Zannella, C.; Carbone, V.; Minasi, P.; Folliero, V.; Stelitano, D.; Cara, F.; Galdiero, M.; Franci, G.; Morana, A. Grape Canes from Typical Cultivars of Campania (Southern Italy) as a Source of High-Value Bioactive Compounds: Phenolic Profile, Antioxidant and Antimicrobial Activities. Molecules 2021, 26, 2746. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, M.; Zhang, L.; Li, L.; Zhu, G.; Shen, E.; Lv, M.; Lu, X.; Sun, Z. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J. Clin. Lab. Anal. 2021, 35, e23655. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zannella, C.; Rinaldi, L.; Boccia, G.; Chianese, A.; Sasso, F.C.; De Caro, F.; Franci, G.; Galdiero, M. Regulation of m6A Methylation as a New Therapeutic Option against COVID-19. Pharmaceuticals 2021, 14, 1135. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14111135
Zannella C, Rinaldi L, Boccia G, Chianese A, Sasso FC, De Caro F, Franci G, Galdiero M. Regulation of m6A Methylation as a New Therapeutic Option against COVID-19. Pharmaceuticals. 2021; 14(11):1135. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14111135
Chicago/Turabian StyleZannella, Carla, Luca Rinaldi, Giovanni Boccia, Annalisa Chianese, Ferdinando Carlo Sasso, Francesco De Caro, Gianluigi Franci, and Massimiliano Galdiero. 2021. "Regulation of m6A Methylation as a New Therapeutic Option against COVID-19" Pharmaceuticals 14, no. 11: 1135. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14111135