PnBA-b-PNIPAM-b-PDMAEA Thermo-Responsive Triblock Terpolymers and Their Quaternized Analogs as Gene and Drug Delivery Vectors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Triblock Terpolymer Synthesis
2.3. Polyplex Formation
2.4. Preparation of CUR-Loaded Micellar Aggregates
2.5. Light Scattering
2.6. UV–Vis
2.7. Fluorescence Spectroscopy
2.7.1. Ethidium Bromide Quenching Assay
2.7.2. Fluorescence of CUR-Loaded Aggregates
3. Results and Discussion
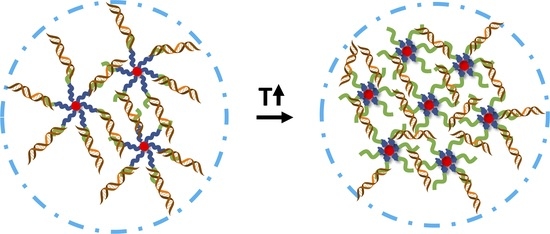
3.1. PDMAEMA-b-PNIPAM-b-(Q)PDMAEA/DNA Polyplexes
3.2. Encapsulation of CUR in the PnBA-b-PNIPAM-b-PDMAEA Polymeric Micellar Aggregates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef]
- Sung, Y.K.; Kim, S.W. Recent Advances in the Development of Gene Delivery Systems. Biomater. Res. 2019, 23, 8. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.J.; Tajmir-Riahi, H.-A.; Pillai, C.K.S. Biodegradable Polymers for Gene Delivery. Molecules 2019, 24, 3744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, W.-F.; Wong, W.-T. Design of Polymeric Gene Carriers for Effective Intracellular Delivery. Trends Biotechnol. 2018, 36, 713–728. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Gersbach, C.A. Engineering Delivery Vehicles for Genome Editing. Annu. Rev. Chem. Biomol. Eng. 2016, 7, 637–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Huang, L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012, 45, 971–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayerossadat, N.; Maedeh, T.; Ali, P.A. Viral and Nonviral Delivery Systems for Gene Delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef]
- Dinçer, S.; Türk, M.; Pişkin, E. Intelligent Polymers as Nonviral Vectors. Gene Ther. 2005, 12, S139–S145. [Google Scholar] [CrossRef] [Green Version]
- Kundu, P.P.; Sharma, V. Synthetic Polymeric Vectors in Gene Therapy. Curr. Opin. Solid State Mater. Sci. 2008, 12, 89–102. [Google Scholar] [CrossRef]
- Pathak, A.; Patnaik, S.; Gupta, K.C. Recent Trends in Non-Viral Vector-Mediated Gene Delivery. Biotechnol. J. 2009, 4, 1559–1572. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Navarro, G.; Pan, J.; Torchilin, V.P. Micelle-like Nanoparticles as Carriers for DNA and SiRNA. Mol. Pharm. 2015, 12, 301–313. [Google Scholar] [CrossRef]
- Zhang, X.-X.; McIntosh, T.J.; Grinstaff, M.W. Functional Lipids and Lipoplexes for Improved Gene Delivery. Biochimie 2012, 94, 42–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.K.; Dang, C.; Wang, X.; Ragan, R.; Kwon, Y.J. Mixing-Sequence-Dependent Nucleic Acid Complexation and Gene Transfer Efficiency by Polyethylenimine. Biomater. Sci. 2015, 3, 1124–1133. [Google Scholar] [CrossRef] [Green Version]
- Byrne, M.; Victory, D.; Hibbitts, A.; Lanigan, M.; Heise, A.; Cryan, S.-A. Molecular Weight and Architectural Dependence of Well-Defined Star-Shaped Poly(Lysine) as a Gene Delivery Vector. Biomater. Sci. 2013, 1, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Dimitrov, I.; Kozarova, R.; Turmanova, S.; Apostolova, M. Thermally Sensitive Polypeptide-Based Copolymer for DNA Complexation into Stable Nanosized Polyplexes. J. Nanopart. Res. 2012, 15, 1358. [Google Scholar] [CrossRef]
- Garrett, S.W.; Davies, O.R.; Milroy, D.A.; Wood, P.J.; Pouton, C.W.; Threadgill, M.D. Synthesis and Characterisation of Polyamine–Poly(Ethylene Glycol) Constructs for DNA Binding and Gene Delivery. Bioorganic Med. Chem. 2000, 8, 1779–1797. [Google Scholar] [CrossRef]
- Nisha, C.K.; Manorama, S.v.; Ganguli, M.; Maiti, S.; Kizhakkedathu, J.N. Complexes of Poly(Ethylene Glycol)-Based Cationic Random Copolymer and Calf Thymus DNA: A Complete Biophysical Characterization. Langmuir 2004, 20, 2386–2396. [Google Scholar] [CrossRef]
- Crommelin, D.J.A.; Storm, G.; Jiskoot, W.; Stenekes, R.; Mastrobattista, E.; Hennink, W.E. Nanotechnological Approaches for the Delivery of Macromolecules. J. Control. Release 2003, 87, 81–88. [Google Scholar] [CrossRef]
- De Smedt, S.C.; Demeester, J.; Hennink, W.E. Cationic Polymer Based Gene Delivery Systems. Pharm. Res. 2000, 17, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, A.; Uchman, M.; Štěpánek, M.; Kereïche, S.; Pispas, S. Complexation of DNA with QPDMAEMA-b-PLMA-b-POEGMA Cationic Triblock Terpolymer Micelles. Macromolecules 2020, 53, 5747–5755. [Google Scholar] [CrossRef]
- Giaouzi, D.; Pispas, S. Complexation Behavior of PNIPAM-b-QPDMAEA Copolymer Aggregates with Linear DNAs of Different Lengths. Eur. Polym. J. 2021, 155, 110575. [Google Scholar] [CrossRef]
- Chroni, A.; Pispas, S. Hydrophilic/Hydrophobic Modifications of a PnBA-b-PDMAEA Copolymer and Complexation Behaviour with Short DNA. Eur. Polym. J. 2020, 129, 109636. [Google Scholar] [CrossRef]
- Shim, M.S.; Kwon, Y.J. Stimuli-Responsive Polymers and Nanomaterials for Gene Delivery and Imaging Applications. Adv. Drug Deliv. Rev. 2012, 64, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Gao, Y.; Li, X.; Serpe, M.J. Stimuli-Responsive Polymers and Their Applications. Polym. Chem. 2017, 8, 127–143. [Google Scholar] [CrossRef] [Green Version]
- Pelton, R. Poly(N-Isopropylacrylamide) (PNIPAM) Is Never Hydrophobic. J. Colloid Interface Sci. 2010, 348, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Halperin, A.; Kröger, M.; Winnik, F.M. Poly(N-Isopropylacrylamide) Phase Diagrams: Fifty Years of Research. Angew. Chem. Int. Ed. 2015, 54, 15342–15367. [Google Scholar] [CrossRef]
- Fliervoet, L.A.L.; van Nostrum, C.F.; Hennink, W.E.; Vermonden, T. Balancing Hydrophobic and Electrostatic Interactions in Thermosensitive Polyplexes for Nucleic Acid Delivery. Multifunct. Mater. 2019, 2, 024002. [Google Scholar] [CrossRef]
- Türk, M.; Dinçer, S.; Yuluğ, I.G.; Pişkin, E. In Vitro Transfection of HeLa Cells with Temperature Sensitive Polycationic Copolymers. J. Control. Release 2004, 96, 325–340. [Google Scholar] [CrossRef] [Green Version]
- Feng, G.; Chen, H.; Li, J.; Huang, Q.; Gupte, M.J.; Liu, H.; Song, Y.; Ge, Z. Gene Therapy for Nucleus Pulposus Regeneration by Heme Oxygenase-1 Plasmid DNA Carried by Mixed Polyplex Micelles with Thermo-Responsive Heterogeneous Coronas. Biomaterials 2015, 52, 1–13. [Google Scholar] [CrossRef]
- Ma, Y.; Hou, S.; Ji, B.; Yao, Y.; Feng, X. A Novel Temperature-Responsive Polymer as a Gene Vector. Macromol. Biosci. 2010, 10, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Türk, M.; Dinçer, S.; Pişkin, E. Smart and Cationic Poly(NIPA)/PEI Block Copolymers as Non-Viral Vectors: In Vitro and in Vivo Transfection Studies. J. Tissue Eng. Regen. Med. 2007, 1, 377–388. [Google Scholar] [CrossRef]
- Mao, Z.; Ma, L.; Yan, J.; Yan, M.; Gao, C.; Shen, J. The Gene Transfection Efficiency of Thermoresponsive N,N,N-Trimethyl Chitosan Chloride-g-Poly(N-Isopropylacrylamide) Copolymer. Biomaterials 2007, 28, 4488–4500. [Google Scholar] [CrossRef]
- Calejo, M.T.; Cardoso, A.M.S.; Kjøniksen, A.-L.; Zhu, K.; Morais, C.M.; Sande, S.A.; Cardoso, A.L.; Lima, M.C.P.d.; Jurado, A.; Nyström, B. Temperature-Responsive Cationic Block Copolymers as Nanocarriers for Gene Delivery. Int. J. Pharm. 2013, 448, 105–114. [Google Scholar] [CrossRef]
- Kanto, R.; Yonenuma, R.; Yamamoto, M.; Furusawa, H.; Yano, S.; Haruki, M.; Mori, H. Mixed Polyplex Micelles with Thermoresponsive and Lysine-Based Zwitterionic Shells Derived from Two Poly(Vinyl Amine)-Based Block Copolymers. Langmuir 2021, 37, 3001–3014. [Google Scholar] [CrossRef]
- Haladjova, E.; Toncheva-Moncheva, N.; Apostolova, M.D.; Trzebicka, B.; Dworak, A.; Petrov, P.; Dimitrov, I.; Rangelov, S.; Tsvetanov, C.B. Polymeric Nanoparticle Engineering: From Temperature-Responsive Polymer Mesoglobules to Gene Delivery Systems. Biomacromolecules 2014, 15, 4377–4395. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Shah, A.; Siddiq, M.; Kraatz, H.-B. Polymeric Micelles as Drug Delivery Vehicles. RSC Adv. 2014, 4, 17028–17038. [Google Scholar] [CrossRef]
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; del Favero, E.; Cantù, L.; Nicoli, S. Polymeric Micelles in Drug Delivery: An Insight of the Techniques for Their Characterization and Assessment in Biorelevant Conditions. J. Control. Release 2021, 332, 312–336. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Y.; Li, S. Polymeric Micelles: Nanocarriers for Cancer-Targeted Drug Delivery. AAPS PharmSciTech 2014, 15, 862–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisht, S.; Feldmann, G.; Soni, S.; Ravi, R.; Karikar, C.; Maitra, A.; Maitra, A. Polymeric Nanoparticle-Encapsulated Curcumin (“nanocurcumin”): A Novel Strategy for Human Cancer Therapy. J. Nanobiotechnol. 2007, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its Effects on Human Health. Foods 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Tomeh, M.; Hadianamrei, R.; Zhao, X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Teng, C.P.; Win, K.Y.; Chen, Y.; Zhang, X.; Yang, D.-P.; Li, Z.; Ye, E. Polymeric Encapsulation of Turmeric Extract for Bioimaging and Antimicrobial Applications. Macromol. Rapid Commun. 2019, 40, 1800216. [Google Scholar] [CrossRef]
- Skandalis, A.; Pispas, S. PH- and Thermo-Responsive Solution Behavior of Amphiphilic, Linear Triblock Terpolymers. Polymer 2018, 157, 9–18. [Google Scholar] [CrossRef]
- Lowry, G.V.; Hill, R.J.; Harper, S.; Rawle, A.F.; Hendren, C.O.; Klaessig, F.; Nobbmann, U.; Sayre, P.; Rumble, J. Guidance to Improve the Scientific Value of Zeta-Potential Measurements in NanoEHS. Environ. Sci. Nano 2016, 3, 953–965. [Google Scholar] [CrossRef]
- Selianitis, D.; Pispas, S. P(MMA-Co-HPMA)-b-POEGMA Copolymers: Synthesis, Micelle Formation in Aqueous Media and Drug Encapsulation. Polym. Int. 2021. [Google Scholar] [CrossRef]












| Sample | Mw a (10−4) (g mol−1) | Mw/Mn a | Composition b (% wt) PnBA/PNIPAM/(Q)PDMAEA |
|---|---|---|---|
| PnBA39-b-PNIPAM87-b-PDMAEA17 | 1.82 | 1.25 | 28/59/13 |
| PnBA39-b-PNIPAM87-b-PDMAEA35 | 2.40 | 1.27 | 25/54/21 |
| PnBA39-b-PNIPAM87-b-QPDMAEA17 | 2.05 c | - | 26/51/23 |
| PnBA39-b-PNIPAM87-b-QPDMAEA35 | 2.90 c | - | 21/45/34 |
| Sample | CUR (%) | Temperature (°C) | Intensity a (a.u.) Free/Loaded | Rh a (nm) Free/Loaded | PDI a Free/Loaded |
|---|---|---|---|---|---|
| PnBA39-b-PNIPAM87-b-PDMAEA35 | 10 | 25 | 5960/14,870 | 83/71 | 0.35/0.16 |
| 45 | 19,000/30,000 | 54/73 | 0.22/0.14 | ||
| 20 | 25 | 5960/17,450 | 83/61 | 0.35/0.13 | |
| 45 | 19,000/37,000 | 54/64 | 0.22/0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skandalis, A.; Selianitis, D.; Pispas, S. PnBA-b-PNIPAM-b-PDMAEA Thermo-Responsive Triblock Terpolymers and Their Quaternized Analogs as Gene and Drug Delivery Vectors. Polymers 2021, 13, 2361. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13142361
Skandalis A, Selianitis D, Pispas S. PnBA-b-PNIPAM-b-PDMAEA Thermo-Responsive Triblock Terpolymers and Their Quaternized Analogs as Gene and Drug Delivery Vectors. Polymers. 2021; 13(14):2361. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13142361
Chicago/Turabian StyleSkandalis, Athanasios, Dimitrios Selianitis, and Stergios Pispas. 2021. "PnBA-b-PNIPAM-b-PDMAEA Thermo-Responsive Triblock Terpolymers and Their Quaternized Analogs as Gene and Drug Delivery Vectors" Polymers 13, no. 14: 2361. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13142361