Reconstruction of Ocean Color Data Using Machine Learning Techniques in Polar Regions: Focusing on Off Cape Hallett, Ross Sea
Abstract
:1. Introduction
2. Study Sites
3. Materials
3.1. Satellite and Reanalysis Data
3.1.1. Sea Surface Temperature
3.1.2. Sea Ice Concentration
3.1.3. Atmospheric Components
3.1.4. Geographical Information
3.1.5. Chlorophyll Concentration
3.2. In-Situ Chlorophyll Concentration
4. Methods
4.1. Predictor Selection
4.2. Data Preprocessing
4.3. Machine Learning Models
4.4. Model Development
4.4.1. Model Comparison
4.4.2. Class Imbalance and Noise on CHL Data
4.4.3. Determination of Hyperparameters
5. Results
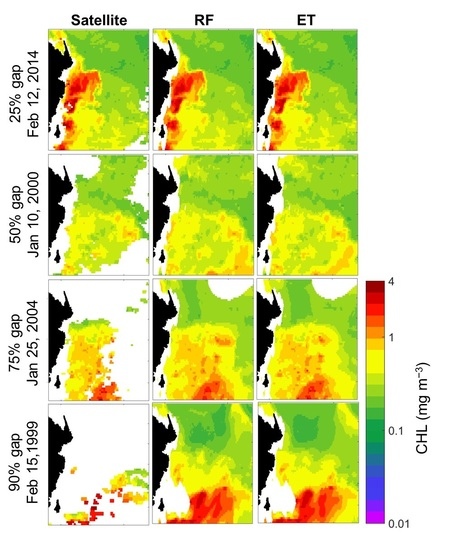
5.1. Reconstruction of CHL Data
5.2. Contribution of Predictor Variables
6. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- O’Reilly, J.E.; Maritorena, S.; Mitchell, B.G.; Siegel, D.A.; Carder, K.L.; Garver, S.A.; Kahru, M.; McClain, C. Ocean color chlorophyll algorithms for SeaWiFS. J. Geophys. Res. Ocean. 1998, 103, 24937–24953. [Google Scholar] [CrossRef] [Green Version]
- Blondeau-Patissier, D.; Gower, J.F.R.; Dekker, A.G.; Phinn, S.R.; Brando, V.E. A review of ocean color remote sensing methods and statistical techniques for the detection, mapping and analysis of phytoplankton blooms in coastal and open oceans. Prog. Oceanogr. 2014, 123, 23–144. [Google Scholar] [CrossRef]
- Klemas, V. Remote Sensing Techniques for Studying Coastal Ecosystems: An Overview. J. Coast. Res. 2011, 27, 2–17. [Google Scholar] [CrossRef]
- Wilson, C. The rocky road from research to operations for satellite ocean-colour data in fishery management. ICES J. Mar. Sci. 2011, 68, 677–686. [Google Scholar] [CrossRef]
- Shen, L.; Xu, H.; Guo, X. Satellite remote sensing of harmful algal blooms (HABs) and a potential synthesized framework. Sensors (Switzerland) 2012, 12, 7778–7803. [Google Scholar] [CrossRef]
- Nechad, B.; Alvera-Azcaràte, A.; Ruddick, K.; Greenwood, N. Reconstruction of MODIS total suspended matter time series maps by DINEOF and validation with autonomous platform data. Ocean Dyn. 2011, 61, 1205–1214. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, M. Filling the Gaps of Missing Data in the Merged VIIRS SNPP/NOAA-20 Ocean Color Product Using the DINEOF Method. Remote Sens. 2019, 11, 178. [Google Scholar] [CrossRef]
- Chen, S.; Hu, C.; Barnes, B.B.; Xie, Y.; Lin, G.; Qiu, Z. Improving ocean color data coverage through machine learning. Remote Sens. Environ. 2019, 222, 286–302. [Google Scholar] [CrossRef]
- Jouini, M.; Lévy, M.; Crépon, M.; Thiria, S. Reconstruction of satellite chlorophyll images under heavy cloud coverage using a neural classification method. Remote Sens. Environ. 2013, 131, 232–246. [Google Scholar] [CrossRef]
- Jo, Y.; Kim, D.; Kim, H. Chlorophyll Concentration Derived from Microwave Remote Sensing Measurements USING Artificial Neural Network Algorithm. J. Mar. Sci 2018, 26, 102–110. [Google Scholar] [CrossRef]
- Arrigo, K.R.; Van Dijken, G.L. Annual changes in sea-ice, chlorophyll a, and primary production in the Ross Sea, Antarctica. Deep. Res. Part II Top. Stud. Oceanogr. 2004, 51, 117–138. [Google Scholar] [CrossRef]
- Marrari, M.; Hu, C.; Daly, K. Validation of SeaWiFS chlorophyll a concentrations in the Southern Ocean: A revisit. Remote Sens. Environ. 2006, 105, 367–375. [Google Scholar] [CrossRef]
- Alvera-Azcárate, A.; Barth, A.; Beckers, J.M.; Weisberg, R.H. Multivariate reconstruction of missing data in sea surface temperature, chlorophyll, and wind satellite fields. J. Geophys. Res. Ocean. 2007, 112. [Google Scholar] [CrossRef] [Green Version]
- Alvera-Azcárate, A.; Barth, A.; Sirjacobs, D.; Beckers, J.M. Enhancing temporal correlations in EOF expansions for the reconstruction of missing data using DINEOF. Ocean Sci. 2009, 5, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Ruddick, K.; Lacroix, G.; Alvera-Azcárate, A.; Park, Y.; Nechad, B.; Sirjacobs, D.; Barth, A.; Beckers, J.-M. Cloud filling of ocean colour and sea surface temperature remote sensing products over the Southern North Sea by the Data Interpolating Empirical Orthogonal Functions methodology. J. Sea Res. 2010, 65, 114–130. [Google Scholar] [CrossRef]
- Volpe, G.; Nardelli, B.B.; Cipollini, P.; Santoleri, R.; Robinson, I.S. Seasonal to interannual phytoplankton response to physical processes in the Mediterranean Sea from satellite observations. Remote Sens. Environ. 2012, 117, 223–235. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, D. Reconstruction of satellite chlorophyll-a data using a modified DINEOF method: A case study in the Bohai and Yellow seas, China. Int. J. Remote Sens. 2014, 35, 204–217. [Google Scholar] [CrossRef]
- Zhao, Y.; He, R. Cloud-free sea surface temperature and colour reconstruction for the gulf of mexico: 2003–2009. Remote Sens. Lett. 2012, 3, 697–706. [Google Scholar] [CrossRef]
- Alvera-Azcárate, A.; Vanhellemont, Q.; Ruddick, K.; Barth, A.; Beckers, J.M. Analysis of high frequency geostationary ocean colour data using DINEOF. Estuar. Coast. Shelf Sci. 2015, 159, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Dreano, D.; Mallick, B.; Hoteit, I. Filtering remotely sensed chlorophyll concentrations in the Red Sea using a space-time covariance model and a Kalman filter. Spat. Stat. 2015, 13, 1–20. [Google Scholar] [CrossRef]
- Hilborn, A.; Costa, M. Applications of DINEOF to Satellite-Derived Chlorophyll-a from a Productive Coastal Region. Remote Sens. 2018, 10, 1449. [Google Scholar] [CrossRef]
- Jayaram, C.; Priyadarshi, N.; Pavan Kumar, J.; Udaya Bhaskar, T.V.S.; Raju, D.; Kochuparampil, A.J. Analysis of gap-free chlorophyll-a data from MODIS in Arabian Sea, reconstructed using DINEOF. Int. J. Remote Sens. 2018, 39, 7506–7522. [Google Scholar] [CrossRef]
- Liu, M.; Liu, X.; Ma, A.; Li, T.; Du, Z. Spatio-temporal stability and abnormality of chlorophyll-a in the northern south china sea during 2002-2012 from MODIS images using wavelet analysis. Cont. Shelf Res. 2014, 75, 15–27. [Google Scholar] [CrossRef]
- Liu, X.; Wang, M. Analysis of ocean diurnal variations from the Korean Geostationary Ocean Color Imager measurements using the DINEOF method. Estuar. Coast. Shelf Sci. 2016, 180, 230–241. [Google Scholar] [CrossRef]
- Liu, X.; Wang, M. Gap filling of missing data for VIIRS global ocean color products using the DINEOF Method. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4464–4476. [Google Scholar] [CrossRef]
- Miles, T.N.; He, R. Temporal and spatial variability of Chl-a and SST on the South Atlantic Bight: Revisiting with cloud-free reconstructions of MODIS satellite imagery. Cont. Shelf Res. 2010, 30, 1951–1962. [Google Scholar] [CrossRef]
- Krasnopolsky, V.; Nadiga, S.; Mehra, A.; Bayler, E.; Behringer, D. Neural networks technique for filling gaps in satellite measurements: Application to ocean color observations. Comput. Intell. Neurosci. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Guisan, A.; Zimmermann, N.E. Predictive habitat distribution models in ecology. Ecol. Modell. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Jane, E.; Catherine, H.G.; Robert, P.A.; Miroslav, D.; Simon, F.; Antoine, G.; Robert, J.H.; Falk, H.; John, R.L.; Anthony, L.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography (Cop.). 2006, 29. [Google Scholar] [CrossRef]
- Smoliński, S.; Radtke, K. Spatial prediction of demersal fish diversity in the Baltic Sea: Comparison of machine learning and regression-based techniques. ICES J. Mar. Sci. 2017, 74, 102–111. [Google Scholar] [CrossRef]
- Beckers, J.M.; Rixen, M. EOF calculations and data filling from incomplete oceanographic datasets. J. Atmos. Ocean. Technol. 2003, 20, 1839–1856. [Google Scholar] [CrossRef]
- Smith, W.O.; Asper, V.L. The influence of phytoplankton assemblage composition on biogeochemical characteristics and cycles in the southern Ross Sea, Antarctica. Deep. Res. Part I Oceanogr. Res. Pap. 2001, 48, 137–161. [Google Scholar] [CrossRef]
- Boyd, P.W. Environmental factors controlling phytoplankton processes in the Southern Ocean. J. Phycol. 2002, 38, 844–861. [Google Scholar] [CrossRef]
- Jones, R.M.; Smith, W.O. The influence of short-term events on the hydrographic and biological structure of the southwestern Ross Sea. J. Mar. Syst. 2017, 166, 184–195. [Google Scholar] [CrossRef] [Green Version]
- Peloquin, J.A.; Smith, W.O. Phytoplankton blooms in the Ross Sea, Antarctica: Interannual variability in magnitude, temporal patterns, and composition. J. Geophys. Res. Ocean. 2007, 112, 1–12. [Google Scholar] [CrossRef]
- Ryan-Keogh, T.J.; DeLizo, L.M.; Smith, W.O.; Sedwick, P.N.; McGillicuddy, D.J.; Moore, C.M.; Bibby, T.S. Temporal progression of photosynthetic-strategy in phytoplankton in the Ross Sea, Antarctica. J. Mar. Syst. 2017, 166, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Sedwick, P.N.; Marsay, C.M.; Sohst, B.M.; Aguilar-Islas, A.M.; Lohan, M.C.; Long, M.C.; Arrigo, K.R.; Dunbar, R.B.; Saito, M.A.; Smith, W.O.; et al. Early season depletion of dissolved iron in the Ross Sea polynya: Implications for iron dynamics on the Antarctic continental shelf. J. Geophys. Res. Ocean. 2011, 116, 1–19. [Google Scholar] [CrossRef]
- Lynch, H.J.; LaRue, M.A. First global census of the Adélie Penguin. Auk Ornithol. Adv. 2014, 131, 457–466. [Google Scholar] [CrossRef]
- Emslie, S.D.; McKenzie, A.; Patterson, W.P. The rise and fall of an ancient adélie penguin ‘supercolony’ at cape adare, antarctica. R. Soc. Open Sci. 2018, 5. [Google Scholar] [CrossRef]
- Weber, L.H.; El-Sayed, S.Z.; Hampton, I. The variance spectra of phytoplankton, krill and water temperature in the Antarctic Ocean south of Africa. Deep Sea Res. Part A, Oceanogr. Res. Pap. 1986, 33, 1327–1343. [Google Scholar] [CrossRef]
- Kaufman, D.E.; Friedrichs, M.A.M.; Smith, W.O.; Queste, B.Y.; Heywood, K.J. Biogeochemical variability in the southern Ross Sea as observed by a glider deployment. Deep. Res. Part I Oceanogr. Res. Pap. 2014, 92, 93–106. [Google Scholar] [CrossRef]
- Lyver, P.O.B.; MacLeod, C.J.; Ballard, G.; Karl, B.J.; Barton, K.J.; Adams, J.; Ainley, D.G.; Wilson, P.R. Intra-seasonal variation in foraging behavior among Adélie penguins (Pygocelis adeliae) breeding at Cape Hallett, Ross Sea, Antarctica. Polar Biol. 2011, 34, 49–67. [Google Scholar] [CrossRef]
- Reynolds, R.W.; Smith, T.M.; Liu, C.; Chelton, D.B.; Casey, K.S.; Schlax, M.G. Daily high-resolution-blended analyses for sea surface temperature. J. Clim. 2007, 20, 5473–5496. [Google Scholar] [CrossRef]
- EUMETSAT Ocean and Sea Ice Satelitte Application Facility. Global sea ice concentration continuous reprocessed product (year), [Online]. Norwegian and Danish Meteorological Institutes. Available online: http://osisaf.met.no (accessed on 5 June 2019).
- Comiso, J.C.; Cavalieri, D.J.; Parkinson, C.L.; Gloersen, P. Passive microwave algorithms for sea ice concentration: A comparison of two techniques. Remote Sens. Environ. 1997, 60, 357–384. [Google Scholar] [CrossRef]
- Dee, D.P.; Uppala, S.M.; Simmons, A.J.; Berrisford, P.; Poli, P.; Kobayashi, S.; Andrae, U.; Balmaseda, M.A.; Balsamo, G.; Bauer, P.; et al. The ERA-Interim reanalysis: Configuration and performance of the data assimilation system. Q. J. R. Meteorol. Soc. 2011, 137, 553–597. [Google Scholar] [CrossRef]
- Fogt, R.L.; Wovrosh, A.J.; Langen, R.A.; Simmonds, I. The characteristic variability and connection to the underlying synoptic activity of the Amundsen-Bellingshausen Seas Low. J. Geophys. Res. Atmos. 2012, 117, 1–22. [Google Scholar] [CrossRef]
- Coggins, J.H.J.; McDonald, A.J.; Jolly, B. Synoptic climatology of the Ross Ice Shelf and Ross Sea region of Antarctica: K-means clustering and validation. Int. J. Climatol. 2014, 34, 2330–2348. [Google Scholar] [CrossRef]
- Dale, E.R.; McDonald, A.J.; Coggins, J.H.J.; Rack, W. Atmospheric forcing of sea ice anomalies in the Ross Sea polynya region. Cryosphere 2017, 11, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Sanz Rodrigo, J.; Buchlin, J.M.; van Beeck, J.; Lenaerts, J.T.M.; van den Broeke, M.R. Evaluation of the antarctic surface wind climate from ERA reanalyses and RACMO2/ANT simulations based on automatic weather stations. Clim. Dyn. 2013, 40, 353–376. [Google Scholar] [CrossRef]
- Gebco Gridded Global Bathymetry Data; British Oceanographic Data Centre: Liverpool, UK, 2009.
- Becker, J.J.; Sandwell, D.T.; Smith, W.H.F.; Braud, J.; Binder, B.; Depner, J.; Fabre, D.; Factor, J.; Ingalls, S.; Kim, S.H.; et al. Global Bathymetry and Elevation Data at 30 Arc Seconds Resolution: SRTM30_PLUS. Mar. Geod. 2009, 32, 355–371. [Google Scholar] [CrossRef]
- GlobColour data (http://globcolour.info) used in this study has been developed, validated, and distributed by ACRI-ST, France.
- Saba, V.S.; Friedrichs, M.A.M.; Antoine, D.; Armstrong, R.A.; Asanuma, I.; Behrenfeld, M.J.; Ciotti, A.M.; Dowell, M.; Hoepffner, N.; Hyde, K.J.W.; et al. An evaluation of ocean color model estimates of marine primary productivity in coastal and pelagic regions across the globe. Biogeosciences 2011, 8, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Parson, T.R.; Maita, Y.; Lalli, C.M. A Manual of Chemical & Biological Methods for Seawater Analysis; Pergamon Press: New York, NY, USA, 2013; ISBN 0080302882. [Google Scholar]
- Morales Maqueda, M.A.; Willmott, A.J.; Biggs, N.R.T. Polynya dynamics: A review of observations and modeling. Rev. Geophys. 2004, 42. [Google Scholar] [CrossRef]
- Nihashi, S.; Ohshima, K.I. Relationship between ice decay and solar heating through open water in the Antarctic sea ice zone. J. Geophys. Res. Ocean. 2001, 106, 16767–16782. [Google Scholar] [CrossRef] [Green Version]
- Coale, K.H.; Wang, X.; Tanner, S.J.; Johnson, K.S. Phytoplankton growth and biological response to iron and zinc addition in the Ross Sea and Antarctic Circumpolar Current along 170°W. Deep Sea Res. Part II Top. Stud. Oceanogr. 2003, 50, 635–653. [Google Scholar] [CrossRef]
- Sedwick, P.N.; Garcia, N.S.; Riseman, S.F.; Marsay, C.M.; DiTullio, G.R. Evidence for high iron requirements of colonial Phaeocystis antarctica at low irradiance. Phaeocystis Major Link Biogeochem. Cycl. Clim. Elem. 2007, 83–97. [Google Scholar] [CrossRef]
- McGillicuddy, D.J.; Sedwick, P.N.; Dinniman, M.S.; Arrigo, K.R.; Bibby, T.S.; Greenan, B.J.W.; Hofmann, E.E.; Klinck, J.M.; Smith, W.O.; Mack, S.L.; et al. Iron supply and demand in an Antarctic shelf ecosystem. Geophys. Res. Lett. 2015, 42, 8088–8097. [Google Scholar] [CrossRef]
- Reddy, T.E.; Arrigo, K.R. Constraints on the extent of the Ross Sea phytoplankton bloom. J. Geophys. Res. Ocean. 2006, 111. [Google Scholar] [CrossRef] [Green Version]
- Arrigo, K.R.; McClain, C.R. Spring phytoplankton production in the western Ross Sea. Science 1994, 266, 261–263. [Google Scholar] [CrossRef]
- Mangoni, O.; Saggiomo, V.; Bolinesi, F.; Margiotta, F.; Budillon, G.; Cotroneo, Y.; Misic, C.; Rivaro, P.; Saggiomo, M. Phytoplankton blooms during austral summer in the Ross Sea, Antarctica: Driving factors and trophic implications. PLoS One 2017, 12, 1–23. [Google Scholar] [CrossRef]
- Kaufman, D.E.; Friedrichs, M.A.M.; Smith, W.O.; Hofmann, E.E.; Dinniman, M.S.; Hemmings, J.C.P. Climate change impacts on southern Ross Sea phytoplankton composition, productivity, and export. J. Geophys. Res. Ocean. 2017, 122, 2339–2359. [Google Scholar] [CrossRef]
- Smith, W.O.; Ainley, D.G.; Cattaneo-Vietti, R. Trophic interactions within the Ross Sea continental shelf ecosystem. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.O.; Jones, R.M. Vertical mixing, critical depths, and phytoplankton growth in the Ross Sea. ICES J. Mar. Sci. 2015, 72, 1952–1960. [Google Scholar] [CrossRef]
- Mastyło, M. Bilinear interpolation theorems and applications. J. Funct. Anal. 2013, 265, 185–207. [Google Scholar] [CrossRef]
- Khoshgoftaar, T.M.; Van Hulse, J.; Napolitano, A. Comparing boosting and bagging techniques with noisy and imbalanced data. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 2011, 41, 552–568. [Google Scholar] [CrossRef]
- Noi, P.T.; Kappas, M. Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using sentinel-2 imagery. Sensors (Switzerland) 2018, 18. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from imbalanced data. Trans. Knowl. Data Eng. 2008, 21, 1263–1284. [Google Scholar]
- Abd Elrahman, S.M.; Abraham, A. A Review of Class Imbalance Problem. J. Netw. Innov. Comput. 2013, 1, 332–340. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 103. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Li, W.; Yan, X.H. Retrieving Temperature Anomaly in the Global Subsurface and Deeper Ocean From Satellite Observations. J. Geophys. Res. Ocean. 2018, 123, 399–410. [Google Scholar] [CrossRef]
- Pinto, A.; Pereira, S.; Rasteiro, D.; Silva, C.A. Hierarchical brain tumour segmentation using extremely randomized trees. Pattern Recognit. 2018, 82, 105–117. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Du, P.; Samat, A.; Waske, B.; Liu, S.; Li, Z. Random Forest and Rotation Forest for fully polarized SAR image classification using polarimetric and spatial features. ISPRS J. Photogramm. Remote Sens. 2015, 105, 38–53. [Google Scholar] [CrossRef]
- Beckers, J.M.; Barth, A.; Alvera-Azcárate, A. DINEOF reconstruction of clouded images including error maps application to the Sea-Surface Temperature around Corsican Island. Ocean Sci. 2006, 2, 183–199. [Google Scholar] [CrossRef]
- Ping, B.; Su, F.; Meng, Y. An improved DINEOF algorithm for filling missing values in spatio-temporal sea surface temperature data. PLoS One 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- García, V.; Sánchez, J.S.; Mollineda, R.A. On the effectiveness of preprocessing methods when dealing with different levels of class imbalance. Knowledge-Based Syst. 2012, 25, 13–21. [Google Scholar] [CrossRef]
- Lu, B.L.; Wang, X.L.; Yang, Y.; Zhao, H. Learning from imbalanced data sets with a Min-Max modular support vector machine. Front. Electr. Electron. Eng. China 2011, 6, 56–71. [Google Scholar] [CrossRef]
- Smith, W.O.; Dinniman, M.S.; Tozzi, S.; DiTullio, G.R.; Mangoni, O.; Modigh, M.; Saggiomo, V. Phytoplankton photosynthetic pigments in the Ross Sea: Patterns and relationships among functional groups. J. Mar. Syst. 2010, 82, 177–185. [Google Scholar] [CrossRef]
- Arrigo, K.R.; Worthen, D.L.; Robinson, D.H. A coupled ocean-ecosystem model of the Ross Sea: 2. Iron regulation of phytoplankton taxonomic variability and primary production. J. Geophys. Res. 2003, 108, 3231. [Google Scholar] [CrossRef]
- Garrison, D.L.; Jeffries, M.O.; Gibson, A.; Coale, S.L.; Neenan, D.; Fritsen, C.; Okolodkov, Y.B.; Gowing, M.M. Development of sea ice microbial communities during autumn ice formation in the Ross Sea. Mar. Ecol. Prog. Ser. 2003, 259, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ji, R.; Edwards, M.; MacKas, D.L.; Runge, J.A.; Thomas, A.C. Marine plankton phenology and life history in a changing climate: Current research and future directions. J. Plankton Res. 2010, 32, 1355–1368. [Google Scholar] [CrossRef] [PubMed]
- Marchese, C.; Albouy, C.; Tremblay, J.-É.; Dumont, D.; D’Ortenzio, F.; Vissault, S.; Bélanger, S. Changes in phytoplankton bloom phenology over the North Water (NOW) polynya: a response to changing environmental conditions. Polar Biol. 2017, 40. [Google Scholar] [CrossRef]
- Guinder, V.; Molinero, J. Climate Change Effects on Marine Phytoplankton. Mar. Ecol. a Chang. World 2014, 68–90. [Google Scholar] [CrossRef]
- Hales, B.; Takahashi, T. High-resolution biogeochemical investigation of the Ross Sea, Antarctica, during the AESOPS (U. S. JGOFS) Program. Global Biogeochem. Cycles 2004, 18. [Google Scholar] [CrossRef] [Green Version]









| Variables | Abbreviation | Unit | Range | Ordinary Res. | Org | |
|---|---|---|---|---|---|---|
| Predictor | Sea ice concentration | SIC | % | 0–15 | 25 km | OSISAF |
| Sea surface temperature | SST | °C | −1.80–1.67 | 25 km | OISST | |
| 10-m zonal wind | U10 | m s−1 | −16.32 to 20.35 | 25 km | ECMWF | |
| 10-m meridional wind | V10 | m s−1 | −13.22 to 27.29 | |||
| 2-m atmospheric temperature | T2M | K | 242.42–277.46 | |||
| Photosynthetically active radiation | PAR | Jm−2 | 8,234.59–659,284.58 | |||
| Bathymetry | DEP | m | −2,503.97 to −8.01 | ~1 km | GEBCO | |
| Longitude | LON | ° E | 168.02–176.98 | 4 km | GlobColour | |
| Latitude | LAT | ° S | 73.98–71.02 | |||
| Target | Chlorophyll-a concentration | CHL | mg m−3 | 4 km | GlobColour | |
| Expedition | Station No. | Sampling date | Coordinates | CHL (mg m−3) | |
|---|---|---|---|---|---|
| Latitude (°S) | Longitude (°E) | ||||
| ANA08C | S02 | 26 February 2018 | 71.6982 | 172.1864 | 0.37 |
| S03 | 26 February 2018 | 71.9401 | 173.9231 | 0.36 | |
| S05 | 26 February 2018 | 72.1635 | 175.5661 | 0.36 | |
| S06 | 27 February 2018 | 72.5345 | 175.0227 | 0.31 | |
| S07 | 27 February 2018 | 72.2915 | 173.3360 | 0.20 | |
| S09 | 27 February 2018 | 72.0398 | 171.6590 | 0.44 | |
| S11 | 28 February 2018 | 72.9870 | 174.3150 | 0.78 | |
| S12 | 28 February 2018 | 72.7577 | 172.6611 | 0.42 | |
| S14 | 28 February 2018 | 72.5958 | 171.4129 | 0.76 | |
| S17 | 28 February 2018 | 73.4209 | 173.0663 | 1.41 | |
| S18 | 01 March 2018 | 73.1927 | 171.9817 | 1.07 | |
| S19 | 01 March 2018 | 73.0632 | 171.0729 | 1.29 | |
| # Training Data: 1,813,449 # Test Data: 604,483 | Evaluation Metrics | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coefficient of Determination (R2) | Mean Absolute Error (MAE) | Root Mean Squared Error (RMSE) | Relative Absolute Error (RAE) | Relative Squared Error (RSE) | Correlation Coefficient (R) | ||||||||
| Dataset | Tr | Te | Tr | Te | Tr | Te | Tr | Te | Tr | Te | Tr | Te | |
| Model | RF | 0.996 | 0.974 | 0.014 | 0.040 | 0.033 | 0.089 | 0.038 | 0.104 | 0.004 | 0.028 | 0.998 | 0.987 |
| ET | 1.000 | 0.984 | 0.000 | 0.028 | 0.000 | 0.068 | 0.000 | 0.073 | 0.000 | 0.016 | 1.000 | 0.992 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Kim, J.-H.; Kim, H.-c.; Kim, B.-K.; Bae, D.; Jo, Y.-H.; Jo, N.; Lee, S.H. Reconstruction of Ocean Color Data Using Machine Learning Techniques in Polar Regions: Focusing on Off Cape Hallett, Ross Sea. Remote Sens. 2019, 11, 1366. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11111366
Park J, Kim J-H, Kim H-c, Kim B-K, Bae D, Jo Y-H, Jo N, Lee SH. Reconstruction of Ocean Color Data Using Machine Learning Techniques in Polar Regions: Focusing on Off Cape Hallett, Ross Sea. Remote Sensing. 2019; 11(11):1366. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11111366
Chicago/Turabian StylePark, Jinku, Jeong-Hoon Kim, Hyun-cheol Kim, Bong-Kuk Kim, Dukwon Bae, Young-Heon Jo, Naeun Jo, and Sang Heon Lee. 2019. "Reconstruction of Ocean Color Data Using Machine Learning Techniques in Polar Regions: Focusing on Off Cape Hallett, Ross Sea" Remote Sensing 11, no. 11: 1366. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11111366