Hyperspectral Unmixing with Gaussian Mixture Model and Spatial Group Sparsity
Abstract
:1. Introduction
2. Related Models
3. GMM Unmixing with Superpixel Segmentation and Spatial Group Sparsity
3.1. Formulation of the Proposed SGSGMM
3.2. Optimization of the Proposed SGSGMM
3.3. Model Selection
3.4. Implementation Issues
| Algorithm 1 Details for unsupervised of SGSGMM |
| Input: Collected mixed pixel matrix ; the parameter of smoothness and spatial sparsity constraint , and the weight value for the distance metric ; Output: The estimated abundance matrix ;
|
| Algorithm 2 Details for supervised of SGSGMM |
| Input: Collected mixed pixel matrix , endmember ; the parameter of smoothness and spatial sparsity constraint , and the weight value for the distance metric ; Output: The estimated abundance matrix ; Preprocessing: 2: (a) Implement PCA and Generate P spatial groups based on SLIC (b) the confidence index by Equations (8) and (10) 4: Take endmember as input, using CVIC to estimate and calculate by standard EM Set as initialization 6: for each superpixel: while not converged do 8: E step: Calculate by Equation (19) M step: Calculate derivatives of by Equations (23)–(25) 10: Update , , and . end while |
4. Experimental Results
4.1. Synthetic Datasets
4.2. Real-Data Experiments
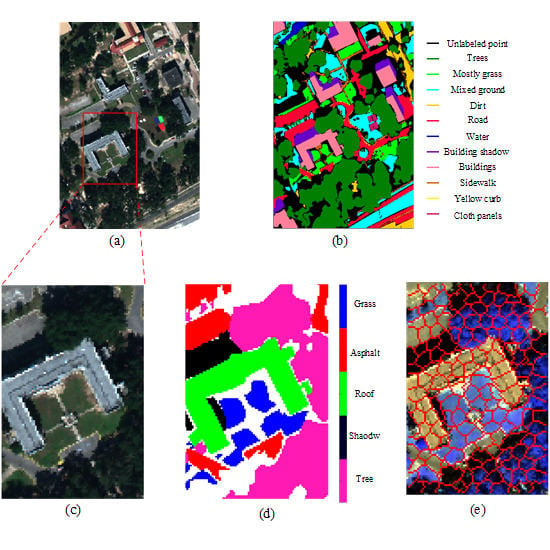
4.2.1. Mississippi Gulfport Datasets
4.2.2. Salinas-A Datasets
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bioucas-Dias, J.M.; Plaza, A.; Dobigeon, N.; Parente, M.; Du, Q.; Gader, P.; Chanussot, J. Hyperspectral unmixing overview: Geometrical, statistical, and sparse regression-based approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 354–379. [Google Scholar] [CrossRef]
- Mei, X.; Ma, Y.; Li, C.; Fan, F.; Huang, J.; Ma, J. Robust GBM hyperspectral image unmixing with superpixel segmentation based low rank and sparse representation. Neurocomputing 2018, 275, 2783–2797. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, J.; Wang, Z.; Chen, C.; Liu, X. Hyperspectral Image Classification in the Presence of Noisy Labels. IEEE Trans. Geosci. Remote Sens. 2019, 57, 851–865. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, H.; Zhao, J.; Gao, Y.; Jiang, J.; Tian, J. Robust feature matching for remote sensing image registration via locally linear transforming. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6469–6481. [Google Scholar] [CrossRef]
- Manolakis, D.; Siracusa, C.; Shaw, G. Hyperspectral subpixel target detection using the linear mixing model. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1392–1409. [Google Scholar] [CrossRef]
- Ma, L.; Crawford, M.M.; Zhu, L.; Liu, Y. Centroid and Covariance Alignment-Based Domain Adaptation for Unsupervised Classification of Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2305–2323. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Jiang, J.; Zhou, H.; Guo, X. Locality preserving matching. Int. J. Comput. Vis. 2019, 127, 512–531. [Google Scholar] [CrossRef]
- Ma, Y.; Jin, Q.; Mei, X.; Dai, X.; Fan, F.; Li, H.; Huang, J. Hyperspectral Unmixing with Gaussian Mixture Model and Low-Rank Representation. Remote Sens. 2019, 11, 911. [Google Scholar] [CrossRef]
- Mei, X.; Pan, E.; Ma, Y.; Dai, X.; Huang, J.; Fan, F.; Du, Q.; Zheng, H.; Ma, J. Spectral-Spatial Attention Networks for Hyperspectral Image Classification. Remote Sens. 2019, 11, 963. [Google Scholar] [CrossRef]
- Li, C.; Ma, Y.; Mei, X.; Fan, F.; Huang, J.; Ma, J. Sparse unmixing of hyperspectral data with noise level estimation. Remote Sens. 2017, 9, 1166. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Xu, H.; Zhang, S.; Mei, X.; Huang, J.; Ma, J. Gaussian field estimator with manifold regularization for retinal image registration. Signal Process. 2019, 157, 225–235. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Luo, L.; Mei, X.; Ma, J. Infrared and visible image fusion based on target-enhanced multiscale transform decomposition. Inf. Sci. 2020, 508, 64–78. [Google Scholar] [CrossRef]
- Fan, F.; Ma, Y.; Li, C.; Mei, X.; Huang, J.; Ma, J. Hyperspectral image denoising with superpixel segmentation and low-rank representation. Inf. Sci. 2017, 397, 48–68. [Google Scholar] [CrossRef]
- Zare, A.; Ho, K. Endmember variability in hyperspectral analysis: Addressing spectral variability during spectral unmixing. IEEE Signal Process. Mag. 2014, 31, 95–104. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P.; Tits, L.; Coppin, P. Endmember variability in spectral mixture analysis: A review. Remote Sens. Environ. 2011, 115, 1603–1616. [Google Scholar] [CrossRef]
- Roberts, D.A.; Gardner, M.; Church, R.; Ustin, S.; Scheer, G.; Green, R. Mapping chaparral in the Santa Monica Mountains using multiple endmember spectral mixture models. Remote Sens. Environ. 1998, 65, 267–279. [Google Scholar] [CrossRef]
- Asner, G.P.; Lobell, D.B. A biogeophysical approach for automated SWIR unmixing of soils and vegetation. Remote Sens. Environ. 2000, 74, 99–112. [Google Scholar] [CrossRef]
- Asner, G.P.; Heidebrecht, K.B. Spectral unmixing of vegetation, soil and dry carbon cover in arid regions: Comparing multispectral and hyperspectral observations. Int. J. Remote Sens. 2002, 23, 3939–3958. [Google Scholar] [CrossRef]
- Dennison, P.E.; Roberts, D.A. Endmember selection for multiple endmember spectral mixture analysis using endmember average RMSE. Remote Sens. Environ. 2003, 87, 123–135. [Google Scholar] [CrossRef]
- Eches, O.; Dobigeon, N.; Mailhes, C.; Tourneret, J.Y. Bayesian estimation of linear mixtures using the normal compositional model. Application to hyperspectral imagery. IEEE Trans. Image Process. 2010, 19, 1403–1413. [Google Scholar] [CrossRef]
- Du, X.; Zare, A.; Gader, P.; Dranishnikov, D. Spatial and spectral unmixing using the beta compositional model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1994–2003. [Google Scholar]
- Zhou, Y.; Rangarajan, A.; Gader, P.D. A Gaussian mixture model representation of endmember variability in hyperspectral unmixing. IEEE Trans. Image Process. 2018, 27, 2242–2256. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Jiang, J.; Liu, C.; Li, Y. Feature guided Gaussian mixture model with semi-supervised EM and local geometric constraint for retinal image registration. Inf. Sci. 2017, 417, 128–142. [Google Scholar] [CrossRef]
- Qu, Q.; Nasrabadi, N.M.; Tran, T.D. Abundance estimation for bilinear mixture models via joint sparse and low-rank representation. IEEE Trans. Geosci. Remote Sens. 2013, 52, 4404–4423. [Google Scholar]
- Eches, O.; Dobigeon, N.; Tourneret, J.Y. Enhancing hyperspectral image unmixing with spatial correlations. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4239. [Google Scholar] [CrossRef]
- Giampouras, P.V.; Themelis, K.E.; Rontogiannis, A.A.; Koutroumbas, K.D. Simultaneously sparse and low-rank abundance matrix estimation for hyperspectral image unmixing. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4775–4789. [Google Scholar] [CrossRef]
- Shi, C.; Wang, L. Incorporating spatial information in spectral unmixing: A review. Remote Sens. Environ. 2014, 149, 70–87. [Google Scholar] [CrossRef]
- Iordache, M.D.; Bioucas-Dias, J.M.; Plaza, A. Total variation spatial regularization for sparse hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2012, 50, 4484–4502. [Google Scholar] [CrossRef]
- Plaza, A.; Martínez, P.; Pérez, R.; Plaza, J. Spatial/spectral endmember extraction by multidimensional morphological operations. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2025–2041. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xia, W.; Wang, B.; Zhang, L. An approach based on constrained non-negative matrix factorization to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2010, 49, 757–772. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, Y.; Zhang, L.; Xu, Y. Spatial group sparsity regularized non-negative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6287–6304. [Google Scholar] [CrossRef]
- Meng, X.L.; Rubin, D.B. Maximum likelihood estimation via the ECM algorithm: A general framework. Biometrika 1993, 80, 267–278. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Lin, Y. Model selection and estimation in regression with grouped variables. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2006, 68, 49–67. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, T. The benefit of group sparsity. Ann. Stat. 2010, 38, 1978–2004. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Huang, Z.; Shen, H.T.; Nie, F. Tag localization with spatial correlations and joint group sparsity. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; pp. 881–888. [Google Scholar]
- Du, Y.; Chang, C.I.; Ren, H.; Chang, C.C.; Jensen, J.O.; D’Amico, F.M. New hyperspectral discrimination measure for spectral characterization. Opt. Eng. 2004, 43, 1777–1787. [Google Scholar]
- Lange, K. The MM algorithm. In Optimization; Springer: New York, NY, USA, 2013; pp. 185–219. [Google Scholar]
- McLachlan, G.J.; Rathnayake, S. On the number of components in a Gaussian mixture model. Wiley Interdiscip. Rev. 2014, 4, 341–355. [Google Scholar] [CrossRef]
- Smyth, P. Model selection for probabilistic clustering using cross-validated likelihood. Stat. Comput. 2000, 10, 63–72. [Google Scholar] [CrossRef]
- Nascimento, J.M.; Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Thompson, D.R.; Mandrake, L.; Gilmore, M.S.; Castano, R. Superpixel endmember detection. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4023–4033. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Nascimento, J.M. Hyperspectral subspace identification. IEEE Trans. Geosci. Remote Sens. 2008, 46, 2435–2445. [Google Scholar] [CrossRef]
- Zhou, Y.; Rangarajan, A.; Gader, P.D. A spatial compositional model for linear unmixing and endmember uncertainty estimation. IEEE Trans. Image Process. 2016, 25, 5987–6002. [Google Scholar] [CrossRef] [PubMed]















| SGSGMM | GMM | SGSNMF | NCM | BCM | |
|---|---|---|---|---|---|
| Asphalt | 208 | 459 | 672 | 566 | 743 |
| Shadow | 80 | 197 | 261 | 278 | 311 |
| Roof | 118 | 340 | 463 | 460 | 586 |
| Grass | 51 | 129 | 175 | 248 | 273 |
| Tree | 81 | 161 | 236 | 262 | 277 |
| Whole map | 107 | 257 | 359 | 363 | 438 |
| SGSGMM | GMM | NCM | |
|---|---|---|---|
| Limestone | 49 | 48 | 51 |
| Basalt | 114 | 138 | 285 |
| Concrete | 90 | 91 | 285 |
| Conifer | 66 | 85 | 81 |
| Asphalt | 115 | 115 | 115 |
| Mean | 87 | 95 | 163 |
| SGSGMM | GMM | SGSNMF | NCM | BCM | |
|---|---|---|---|---|---|
| Asphalt | 189 | 384 | 513 | 474 | 440 |
| Roof | 220 | 333 | 286 | 647 | 660 |
| Grass | 57 | 67 | 95 | 183 | 130 |
| Shadow | 163 | 154 | 158 | 137 | 110 |
| Tree | 385 | 628 | 636 | 767 | 728 |
| Whole map | 158 | 276 | 280 | 357 | 351 |
| SGSGMM | GMM | NCM | |
|---|---|---|---|
| Asphalt | 100 | 232 | 178 |
| Roof | 72 | 227 | 305 |
| Grass | 41 | 71 | 134 |
| Shaodw | 219 | 219 | 566 |
| Tree | 54 | 90 | 112 |
| Mean | 97 | 168 | 259 |
| SGSGMM | GMM | SGSNMF | NCM | BCM | |
|---|---|---|---|---|---|
| Brocoli | 528 | 715 | 511 | 1421 | 511 |
| Corn | 1291 | 2087 | 8068 | 8790 | 8021 |
| Lettuce 4wk | 150 | 2096 | 2766 | 2732 | 2396 |
| Lettuce 5wk | 556 | 520 | 324 | 1858 | 1536 |
| Lettuce 6wk | 530 | 1975 | 9985 | 2529 | 1597 |
| Lettuce 7wk | 790 | 1046 | 1427 | 3053 | 2423 |
| Whole map | 407 | 802 | 2502 | 2268 | 2006 |
| SGSGMM | GMM | NCM | |
|---|---|---|---|
| Brocoli | 315 | 291 | 317 |
| Corn | 140 | 382 | 586 |
| Lettuce 4wk | 177 | 172 | 196 |
| Lettuce 5wk | 63 | 150 | 150 |
| Lettuce 6wk | 116 | 233 | 134 |
| Lettuce 7wk | 151 | 163 | 215 |
| Mean | 160 | 231 | 266 |
| K-Means | VCA | Region-Based VCA | NCM | BCM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SGSGMM | GMM | SGSNMF | SGSGMM | GMM | SGSNMF | SGSGMM | GMM | SGSNMF | |||
| Limestone | 402 | 615 | 818 | 278 | 524 | 668 | 208 | 459 | 672 | 566 | 743 |
| Basalt | 97 | 194 | 330 | 85 | 145 | 262 | 80 | 197 | 261 | 278 | 311 |
| Concrete | 301 | 515 | 533 | 158 | 425 | 448 | 118 | 340 | 463 | 460 | 586 |
| Conifer | 159 | 147 | 254 | 72 | 147 | 173 | 51 | 129 | 175 | 248 | 273 |
| Asphalt | 84 | 190 | 319 | 108 | 192 | 236 | 81 | 161 | 236 | 262 | 277 |
| Whole map | 209 | 332 | 451 | 140 | 287 | 357 | 107 | 257 | 359 | 363 | 438 |
| SGSGMM | GMM | SGSNMF | NCM | BCM | |
|---|---|---|---|---|---|
| Gulfport | 1611 | 2293 | 38 | 268 | 1008 |
| Salinas-A | 803 | 980 | 15 | 528 | 2017 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Q.; Ma, Y.; Pan, E.; Fan, F.; Huang, J.; Li, H.; Sui, C.; Mei, X. Hyperspectral Unmixing with Gaussian Mixture Model and Spatial Group Sparsity. Remote Sens. 2019, 11, 2434. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11202434
Jin Q, Ma Y, Pan E, Fan F, Huang J, Li H, Sui C, Mei X. Hyperspectral Unmixing with Gaussian Mixture Model and Spatial Group Sparsity. Remote Sensing. 2019; 11(20):2434. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11202434
Chicago/Turabian StyleJin, Qiwen, Yong Ma, Erting Pan, Fan Fan, Jun Huang, Hao Li, Chenhong Sui, and Xiaoguang Mei. 2019. "Hyperspectral Unmixing with Gaussian Mixture Model and Spatial Group Sparsity" Remote Sensing 11, no. 20: 2434. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11202434