Assessing the Potential Replacement of Laurel Forest by a Novel Ecosystem in the Steep Terrain of an Oceanic Island
Abstract
:1. Introduction
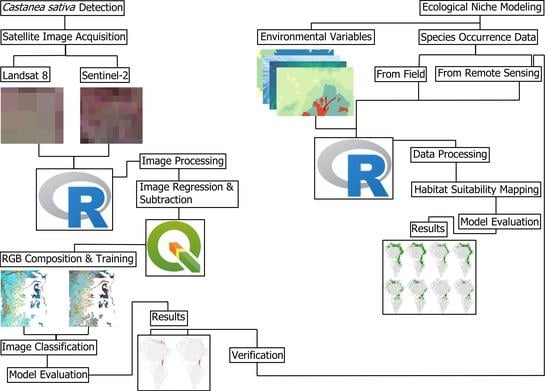
2. Materials and Methods
2.1. Study Site and Field Data
2.2. Change Detection
2.3. Random Forest Classifications
2.4. Ecological Niche Modelling
3. Results
3.1. Change Detection
3.2. Ecological Niche Modelling
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Sensor | Scene Id |
|---|---|
| Landsat 8 | LC082080402017072901T1-SC20190612132658 |
| Landsat 8 | LC082080402017030701T1-SC20190128221754 |
| Landsat 8 | LC082080402017020301T1-SC20190612132509 |
| Sentinel-2 | S2A_MSIL2A_20180708T120331_N0208_R023_T28RBS_20180708T141805 |
| Sentinel-2 | S2A_MSIL2A_20190213T120321_N0211_R023_T28RBS_20190213T172742 |
Appendix B
| Sensor | Data | |
|---|---|---|
| Training | Testing | |
| Sentinel-2 | 101501 | 43499 |
| Landsat 8 | 11557 | 4952 |
Appendix C

Appendix D


Appendix E

Appendix F

Appendix G


Appendix H

Appendix I
| Model | Parameters | |||||
|---|---|---|---|---|---|---|
| AUC | Threshold | TSS | ||||
| Field | RS | Field | RS | Field | RS | |
| EM | 0.982 | 0.961 | 564.833 | 634.417 | 0.885 | 0.805 |
| GAM | NA | 0.943 | NA | 572.540 | NA | 0.789 |
| RF | 0.982 | 0.968 | 570.875 | 675.625 | 0.881 | 0.811 |
| MaxEnt | NA | 0.928 | NA | 634.167 | NA | 0.730 |
References
- Kier, G.; Kreft, H.; Lee, T.M.; Jetz, W.; Ibisch, P.L.; Nowicki, C.; Mutke, J.; Barthlott, W. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. USA 2009, 106, 9322–9327. [Google Scholar] [CrossRef] [Green Version]
- Kreft, H.; Jetz, W.; Mutke, J.; Kier, G.; Barthlott, W. Global diversity of island floras from a macroecological perspective. Ecol. Lett. 2008, 11, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Irl, S.; Schweiger, A.; Medina, F.M.; Fernández-Palacios, J.M.; Harter, D.; Jentsch, A.; Provenzale, A.; Steinbauer, M.J.; Beierkuhnlein, C. An island view of endemic rarity–environmental drivers and consequences for nature conservation. Divers. Distrib. 2017, 23, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Losos, J.B.; Ricklefs, R.E. Adaptation and diversification on islands. Nature 2009, 457, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, A.; White, P.S. A Theory of pulse dynamics and disturbance in ecology. Ecology 2019, 100, e02734. [Google Scholar] [CrossRef]
- Nürk, M.N.; Linder, H.P.; Onstein, R.E.; Marcombe, M.J.; Hughes, C.E.; Pineiro Fernandez, L.; Schlüter, P.M.; Valente, L.M.; Beierkuhnlein, C.; Cutts, V.; et al. Diversification in evolutionary arenas—Assessment and synthesis. Ecol. Evol. 2020, 10, 6163–6182. [Google Scholar] [CrossRef]
- Fernández-Palacios, J.M.; Whittaker, R.J. The Canaries. An important biogeographical meeting place. J. Biogeogr. 2008, 35, 379–387. [Google Scholar] [CrossRef]
- MEA. Ecosystems and Human Well-Beings: Synthesis; Island Press: Washington, DC, USA, 2005. [Google Scholar]
- Pyšek, P.; Jarosik, V.; Hulme, P.E.; Hejda, M.; Schaffner, U.; Vila, M.A. Global assessment of invasive plant impacts on resident species, communities and ecosystems: The interaction of impact measures, invading species traits and environment. Glob. Chang. Biol. 2011, 18, 1725–1737. [Google Scholar] [CrossRef]
- IPBES. Summary for Policymakers of the Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services: Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (Seventh Conference). 2019. Available online: https://www.ipbes.net/system/tdf/ipbes_7_10_add.1_en_1.pdf?file=1&type=node&id=35329 (accessed on 15 September 2020).
- Pyšek, P.; Hulme, P.E.; Simberloff, D.; Bacher, S.; Blackburn, T.M.; Carlton, J.T.; Dawson, W.; Essl, F.; Foxcroft, L.C.; Genovesi, P.; et al. Scientists’ warning on invasive alien species. Biol. Rev. Camb. Philos. Soc. 2020, 95, 1511–1534. [Google Scholar] [CrossRef]
- Mack, R.N.; Simberloff, D.; Lonsdale, W.M.; Evans, H.; Clout, M.; Bazzaz, F.A. Biotic invasions: Causes, epidemiology, global consequences, and control. Ecol. Appl. 2000, 10, 689–710. [Google Scholar] [CrossRef]
- Kumar Rai, P.; Singh, J.S. Invasive alien plant species: Their impact on environment, ecosystem services and human health. Ecol. Indic. 2020, 111, 106020. [Google Scholar] [CrossRef] [PubMed]
- Marbuah, G.; Gren, I.-M.; McKie, B. Economics of harmful invasive species: A review. Diversity 2014, 6, 500–523. [Google Scholar] [CrossRef] [Green Version]
- Díaz, S.; Settele, J.; Brondízio, E.S.; Ngo, H.T.; Agard, J.; Arneth, A.; Balvanera, P.; Brauman, K.A.; Butchart, S.H.M.; Chan, K.M.A.; et al. Pervasive human-driven decline of life on earth points to the need for transformative change. Science 2019, 366, eaax3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oumar, Z. Assessing the utility of the SPOT 6 sensor in detecting and mapping Lantana camara for a community clearing project in KwaZulu-Natal, South Africa. South Afr. J. Geomat. 2016, 5, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Tarantino, C.; Casella, F.; Adamo, M.; Lucas, R.; Beierkuhnlein, C.; Blonda, P. Ailanthus altissima mapping from multi-temporal very high resolution satellite images. J. Photogramm. Remote Sens. 2019, 147, 90–103. [Google Scholar] [CrossRef]
- Weber, E. Invasive Plant Species of the World: A Reference Guide to Environmental Weeds; CAB International Publishing: Wallingford, UK, 2003. [Google Scholar]
- Richardson, D.M.; Rejmánek, M. Conifers as invasive aliens: A global survey and predictive framework. Divers. Distrib. 2004, 10, 321–331. [Google Scholar] [CrossRef]
- Conedera, M.; Krebs, P.; Tinner, W.; Pradella, M.; Torriani, D. The cultivation of Castanea sativa (Mill.) in Europe, form its origin to its diffusion on a continental scale. Veg. Hist. Archaeobotany 2004, 13, 161–179. [Google Scholar] [CrossRef] [Green Version]
- Ríos-Mesa, D.; Pereira-Lorenzo, S.; González-Díaz, A.; Hernádez-González, J.; González-Diaz, E.; Saúco, V.G. The status of Chestnut cultivation and utilization in the Canary Islands. Adv. Hortic. Sci. 2011, 25, 90–98. [Google Scholar]
- Campbell, J.B.; Wynne, H.R. Introduction to Remote Sensing, 5th ed.; The Guilford Press: New York, NY, USA, 2011. [Google Scholar]
- Cohen, W.B.; Goward, S.N. Landsat’s role in ecological applications of remote sensing. BioScience 2004, 54, 535–545. [Google Scholar] [CrossRef]
- Kerr, J.T.; Ostrovsky, M. From space to species: Ecological applications for remote sensing. Trends Ecol. Evol. 2003, 18, 299–305. [Google Scholar] [CrossRef]
- Matongera, T.N.; Mutanga, O.; Dube, T.; Mbulisi, S. Detection and mapping the spatial distribution of bracken fern weeds using the Landsat 8 OLI new generation sensor. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 93–103. [Google Scholar] [CrossRef]
- Hoffmann, S.; Schmitt, T.M.; Chiarucci, A.; Irl, S.D.H.; Rocchini, D.; Vetaas, O.R.; Tasase, M.A.; Mermoz, S.; Bouvet, A.; Beierkuhnlein, C. Remote sensing of β-diversity: Evidence from plant communities in a semi-natural system. Appl. Veg. Sci. 2019, 22, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Vaz, A.S.; Alcaraz-Segura, D.; Vicente, J.R.; Honrado, J.P. The many roles of remote sensing in invasion science. Front. Ecol. Evol. 2019, 7, 370. [Google Scholar] [CrossRef] [Green Version]
- Bolch, E.A.; Santos, M.J.; Ade, C.; Khanna, S.; Basinger, N.T.; Reader, M.O.; Hestir, E.L. Remote detection of invasive alien species. In Remote Sensing of Plant Biodiversity; Cavender-Bares, J., Gamon, J., Townsend, P., Eds.; Springer: Cham, Germany, 2020; pp. 267–307. [Google Scholar]
- Huang, C.Y.; Asner, G.P. Applications of remote sensing to alien invasive plant studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef] [Green Version]
- Rocchini, D.; Andreo, V.; Förster, M.; Garzon-Lopez, C.X.; Gutierrez, A.P.; Gillespie, T.W.; Hauffe, H.C.; He, K.S.; Kleinschmit, B.; Mairota, P.; et al. Potential of remote sensing to predict species invasions. Prog. Phys. Geogr. Earth Environ. 2015, 39, 283–309. [Google Scholar] [CrossRef] [Green Version]
- Evangelista, P.H.; Stohlgren, T.J.; Morisette, J.T.; Kumar, S. Mapping invasive Tamarisk (Tamarix): A comparison of single-scene and time-series analyses of remotely sensed data. Remote Sens. 2009, 1, 519–533. [Google Scholar] [CrossRef] [Green Version]
- Glenn, N.F.; Mundt, J.T.; Weber, K.T.; Prather, T.S.; Lass, L.W.; Pettingill, J. Hyperspectral data processing for repeat detection of small infestations of leafy spurge. Remote Sens. Environ. 2005, 95, 399–412. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Carlson, K.M.; Rascher, U.; Vitousek, P.M. Vegetation–climate interactions among native and invasive species in Hawaiian rainforest. Ecosystems 2006, 9, 1106–1117. [Google Scholar] [CrossRef]
- He, K.S.; Rocchini, D.; Neteler, M.; Nagendra, H. Benefits of hyperspectral remote sensing for tracking plant invasions. Divers. Distrib. 2011, 17, 381–392. [Google Scholar] [CrossRef]
- Vicente, J.R.; Alagador, D.; Guerra, C.; Alonso, J.M.; Kueffer, C.; Vaz, A.S.; Fernandes, R.F.; Cabral, J.A.; Araújo, M.B.; Honrado, J.P. Cost-effective monitoring of biological invasions under global change: A model-based framework. J. Appl. Ecol. 2016, 53, 1317–1329. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Andrew, M.E.; Ustin, S.L. Spectral and physiological uniqueness of perennial pepperweed (Lepidium latifolium). Weed Sci. 2006, 54, 1051–1062. [Google Scholar] [CrossRef]
- Le Louarn, M.; Clergeau, P.; Briche, E.; Deschamps-Cottin, M. “Kill Two Birds with One Stone”: Urban tree species classification using bi-temporal Pléiades images to study nesting preferences of an invasive bird. Remote Sens. 2017, 9, 916. [Google Scholar] [CrossRef] [Green Version]
- Singh, A. Review article digital change detection techniques using remotely sensed data. Int. J. Remote Sens. 1989, 10, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Mai, D.H. Development and regional differentiation of the European vegetation during the Tertiary. In Woody Plants—Evolution and Distribution since the Tertiary; Ehrendorfer, F., Ed.; Springer: Vienna, Austria, 1989. [Google Scholar] [CrossRef]
- Kondraskov, P.; Schütz, N.; Schüßler, C.; De Sequeira, M.M.; Guerra, A.S.; Caujapé-Castells, J.; Jaén-Molina, R.; Marrero-Rodríguez, Á.; Koch, M.A.; Linder, P.; et al. Biogeography of Mediterranean Hotspot Biodiversity: Re-Evaluating the ’Tertiary Relict’ Hypothesis of Macaronesian Laurel Forests. PLoS ONE 2015, 10, e0132091. [Google Scholar] [CrossRef]
- Fernández-Palacios, J.M.; de Nascimento, L.; Otto, R.; Delgado, J.D.; García-del-Rey, E.; Arévalo, J.R.; Whittaker, R.J. A reconstruction of Palaeo-Macaronesia, with particular reference to the long-term biogeography of the Atlantic island laurel forests. J. Biogeogr. 2011, 38, 226–246. [Google Scholar] [CrossRef]
- Fernandez-Palacios, J.M.; Arevalo, J.R.; Balguerias, E.; Barone, R.; de Nascimento, L.; Delgado, J.D.; Elias, R.B.; Fernandez-Lugo, S.; Mendez, J.; de Sequeira, M.M.; et al. The Laurisilva Canaries, Madeira and Azores; Macaronesia Editorial: La Laguna, Spain, 2019. [Google Scholar]
- U.S. Geological Survey. Landsat Surface Reflectance Data; U.S. Geological Survey: Reston, VA, USA, 2015.
- SUHET Sentinel-2 User Handbook (Issue 1.2). Available online: https://sentinels.copernicus.eu/web/sentinel/user-guides/document-library/asset_publisher/xlslt4309D5h/content/sentinel-2-user-handbook (accessed on 5 April 2019).
- Leutner, B.; Horning, N.; Schwalb-Willmann, J. RStoolbox: Tools for Remote Sensing Data Analysis. R Package Version 0.2.6. 2019. Available online: https://CRAN.R-project.org/package=RStoolbox (accessed on 15 September 2020).
- Devi, R.N.; Jiji, D.G. Change detection techniques–A survey. Int. J. Comput. Sci. Eng. 2015, 5, 45–57. [Google Scholar] [CrossRef]
- Tian, S.; Zhang, X.; Tian, J.; Sun, Q. Random forest classification of wetland landcovers from multi-sensor data in the arid region of Xinjiang, China. Remote Sens. 2016, 8, 954. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-86. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 9 September 2020).
- Irl, S.D.H.; Harter, D.E.V.; Steinbauer, M.J.; Puyol, D.G.; Fernández-Palacios, J.M.; Jentsch, A.; Beierkuhnlein, C. Climate vs. topography–Spatial patterns of plant species diversity and endemism on a high-elevation island. J. Ecol. 2015, 103, 1621–1633. [Google Scholar] [CrossRef]
- Cabildo Insular de La Palma. Personal communication, 2019.
- R Core Team. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria. 2020. Available online: https://www.R-project.org/ (accessed on 15 September 2020).
- Thuiller, W.; Georges, D.; Engler, R.; Breiner, F. biomod2: Ensemble Platform for Species Distribution Modeling. R Package Version 3.4.6. 2020. Available online: https://CRAN.R-project.org/package=biomod2 (accessed on 15 September 2020).
- Yee, T.W.; Mitchell, N.D. Generalized additive models in plant ecology. J. Veg. Sci. 1991, 2, 587–602. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Phillis, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Chalghaf, B.; Chemkhi, J.; Mayala, B.; Harrabi, M.; Benie, G.B.; Michael, E.; Salah, A.B. Ecological niche modeling predicting the potential distribution of Leishmania vectors in the Mediterranean basin: Impact of climate change. Parasites Vectors 2018, 11, 461. [Google Scholar] [CrossRef] [PubMed]
- Valvi, R.; Shafizadeh-Moghadam, H.; Matkan, A.; Shakiba, A.; Mirbagheri, B.; Kia, S.H. Modelling climate change effects on Zagros forests in Iran using individual and ensemble forecasting approaches. Theor. Appl. Climatol. 2019, 137, 1015–1025. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.D.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Andrson, R.P. SpThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Manel, S.; Williams, H.C.; Ormerod, S.J. Evaluating presence-absence models in ecology: The need to account for prevalence. J. Appl. Ecol. 2001, 38, 921–931. [Google Scholar] [CrossRef]
- Liu, C.; Berry, P.M.; Dawson, T.P.; Pearson, R.G. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 2005, 28, 385–393. [Google Scholar] [CrossRef]
- Del Arco Aguilar, M.; Wildpret, W.; Pérez de Paz, P.L.; Rodríguez Delgado, O.; Acebes, J.R.; García Gallo, A.; Martín Osorio, V.E.; Reyes Betancort, A.; Salas, M.; Díaz, M.A.; et al. Vegetation Map of the Canary Islands; GRAFCAN: Santa Cruz de Tenerife, Spain, 2006. (In Spanish) [Google Scholar]
- Walentowitz, A.J.; Irl, S.D.H.; Acevedo Rodriguez, A.J.; Palomarez-Martinez, A.; Vetter, V.; Zennaro, B.; Medina, F.M.; Beierkuhnlein, C. Graminoid invasion in an insular endemism hotspot and its protected areas. Diversity 2019, 11, 192. [Google Scholar] [CrossRef] [Green Version]
- Labonté, J.; Drolet, G.; Sylvain, J.-D.; Thiffault, N.; Hébert, F.; Girard, F. Phenology-based mapping of an alien invasive species using time series of multispectral satellite data: A case-study with glossy buckthorn in Québec, Canada. Remote Sens. 2020, 12, 922. [Google Scholar] [CrossRef] [Green Version]
- Price, J.C. How unique are spectral signatures? Remote Sens. Environ. 1994, 49, 181–186. [Google Scholar] [CrossRef]
- Fisher, J.R.B.; Acosta, E.A.; Dennedy-Frank, P.J.; Kroeger, T.; Boucher, T.M. Impact of satellite imagery spatial resolution on land use classification accuracy and modeled water quality. Remote Sens. Ecol. Conserv. 2018, 4, 137–149. [Google Scholar] [CrossRef]
- Arenas-Castro, S.; Fernández-Haeger, J.; Jordano-Barbudo, D. Evaluation and comparison of QuickBird and ADS40-SH52 multispectral imagery for mapping Iberian wild pear trees (Pyrus bourgaeana, Decne) in a Mediterranean mixed forest. Forests 2014, 5, 1304–1330. [Google Scholar] [CrossRef]
- Kennedy, A.T.; Naeem, S.; Howe, K.M.; Knops, J.M.H.; David, T.M.; Peter, R. Biodiversity as a barrier to ecological invasion. Nature 2002, 417, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Stachowicz, J.; Tilman, D. Species Invasions and the Relationships between Species Diversity, Community Saturation, and Ecosystem Functioning. In Species Invasions: Insights into Ecology, Evolution, and Biogeography; Sax, D.F., Stachowicz, J.J., Gaines, S.D., Eds.; Sinauer Associates: Sunderland, MA, USA, 2005; pp. 41–64. [Google Scholar]
- Mallon, C.A.; Poly, F.; Le Roux, X.; Marring, I.; van Elsas, J.D.; Salles, J.F. Resource pulses can alleviate the biodiversity–invasion relationship in soil microbial communities. Ecology 2015, 96, 915–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sax, D.; Gaines, S.D. Species invasions and extinction: The future of native biodiversity on islands. Proc. Natl. Acad. Sci. USA 2005, 105 (Suppl. 1), 11490–11497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateo, R.G.; Mokany, K.; Guisan, A. Biodiversity models: What if unsaturation is the rule? Trends Ecol. Evol. 2017, 32, 556–566. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, R.J.; Higgs, E.; Harris, J.A. Novel ecosystems: Implications for conservation and restoration. Trends Ecol. Evol. 2009, 24, 599–605. [Google Scholar] [CrossRef]
- Carter, G.; Lucas, K.; Blossom, G.; Lassitter, C.; Holiday, D.; Mooneyhan, D.; Fastring, D.; Holcombe, T.; Griffith, J. Remote sensing and mapping of tamarisk along the Colorado River, USA: A comparative use of summer-acquired Hyperion, Thematic Mapper and QuickBird data. Remote Sens. 2009, 1, 318–329. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. Invasive species spread mapping using multi-resolution remote sensing data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. Beijing 2008, 37, 135–142. [Google Scholar]
- Müllerová, J.; Pergl, J.; Pyšek, P. Remote sensing as a tool for monitoring plant invasions: Testing the effects of data resolution and image classification approach on the detection of a model plant species Heracleum mantegazzianum (Giant Hogweed). Int. J. Appl. Earth Obs. Geoinformatics 2013, 25, 55–65. [Google Scholar] [CrossRef]
- Underwood, E.C.; Ustin, S.L.; Ramirez, C.M.A. Comparison of spatial and spectral image resolution for mapping invasive plants in coastal California. Environ. Manag. 2007, 39, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Noujdina, N.V.; Ustin, S.L. Mapping downy brome (Bromus tectorum) using multidate AVIRIS data. Weed Sci. 2008, 56, 173–179. [Google Scholar] [CrossRef]
- Hestir, E.L.; Khanna, S.; Andrew, M.E.; Santos, M.J.; Viers, J.H.; Greenberg, J.A.; Rajapakse, S.S.; Ustin, S.L. Identification of invasive vegetation using hyperspectral remote sensing in the California delta ecosystem. Remote Sens. Environ. 2008, 112, 4034–4047. [Google Scholar] [CrossRef]
- Bradley, B.A.; Mustard, J.F. Characterizing the landscape dynamics of an invasive plant and risk of invasion using remote sensing. Ecol. Appl. 2006, 16, 1132–1147. [Google Scholar] [CrossRef] [Green Version]
- Becker, R.H.; Zmijewski, K.A.; Crail, T. Seeing the forest for the invasives: Mapping buckthorn in the oak openings. Biol. Invasions 2013, 15, 315–326. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty-five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Ullah, S.; Schlerf, M.; Skidmore, A.K.; Hecker, C. Identifying plant species using mid-wave infrared (2.5–6 μm) and thermal infrared (8–14 μm) emissivity spectra. Remote Sens. Environ. 2012, 118, 95–102. [Google Scholar] [CrossRef]
- Stohlgren, T.J.; Ma, P.; Kumar, S.; Rocca, M.; Morisette, J.T.; Jarnevich, C.S.; Benson, N. Ensemble habitat mapping of invasive plant species. Risk Anal. 2010, 30, 224–235. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, O.J.; Sang, W.; Li, Z.; Ma, K. Predicting the spatial distribution of an invasive plant species (Eupatorium adenophorum) in China. Landsc. Ecol. 2007, 22, 1143–1154. [Google Scholar] [CrossRef]
- Warren, D.L. In defense of ‘Niche Modeling’. Trends Ecol. Evol. 2012, 27, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Dehnen-Schmutz, K.; Boivin, T.; Essl, F.; Groom, Q.J.; Harrison, L.; Touza, J.M.; Bayliss, H. Alien futures: What is on the horizon for biological invasions? Divers. Distrib. 2018, 24, 1149–1157. [Google Scholar] [CrossRef] [Green Version]
- Bradley, B.A. Distribution models of invasive plants over-estimate potential impact. Biol. Invasions 2013, 15, 1417–1429. [Google Scholar] [CrossRef]
- Gutierrez, A.P.; Ponti, L. Eradication of invasive species: Why the biology matters. Environ. Entomol. 2013, 42, 395–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feilhauer, H.; Schmidtlein, S. On variable relations between vegetation patterns and canopy reflectance. Ecol. Inform. 2011, 6, 83–92. [Google Scholar] [CrossRef]








| Sensors | Parameters | ||
|---|---|---|---|
| OOB Error % | Overall Accuracy % | Kappa | |
| Landsat 8 | 1.29 | 98.8 | 0.798 |
| Sentinel-2 | 0.44 | 99.5 | 0.878 |
| Model | Parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| AUC | Threshold | TSS | Area (km2) | |||||
| Field | RS | Field | RS | Field | RS | Field | RS | |
| EM | 0.982 | 0.961 | 564.83 | 634.42 | 0.885 | 0.805 | 66.73 | 90.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devkota, R.S.; Field, R.; Hoffmann, S.; Walentowitz, A.; Medina, F.M.; Vetaas, O.R.; Chiarucci, A.; Weiser, F.; Jentsch, A.; Beierkuhnlein, C. Assessing the Potential Replacement of Laurel Forest by a Novel Ecosystem in the Steep Terrain of an Oceanic Island. Remote Sens. 2020, 12, 4013. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12244013
Devkota RS, Field R, Hoffmann S, Walentowitz A, Medina FM, Vetaas OR, Chiarucci A, Weiser F, Jentsch A, Beierkuhnlein C. Assessing the Potential Replacement of Laurel Forest by a Novel Ecosystem in the Steep Terrain of an Oceanic Island. Remote Sensing. 2020; 12(24):4013. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12244013
Chicago/Turabian StyleDevkota, Ram Sharan, Richard Field, Samuel Hoffmann, Anna Walentowitz, Félix Manuel Medina, Ole Reidar Vetaas, Alessandro Chiarucci, Frank Weiser, Anke Jentsch, and Carl Beierkuhnlein. 2020. "Assessing the Potential Replacement of Laurel Forest by a Novel Ecosystem in the Steep Terrain of an Oceanic Island" Remote Sensing 12, no. 24: 4013. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12244013