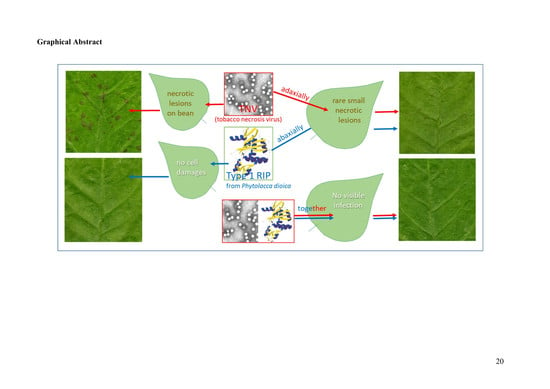
Antiviral Activity of PD-L1 and PD-L4, Type 1 Ribosome Inactivating Proteins from Leaves of Phytolacca dioica L. in the Pathosystem Phaseolus vulgaris–Tobacco Necrosis Virus (TNV)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Type-1 RIP Purification
2.2. Antiviral Activity of PD-L1 and PD-L4
2.3. PD-L1 and PD-L4 Antiviral Activity is not Mediated by Cell Death and Oxidative Burst
3. Conclusions
4. Material and Methods
4.1. Materials
4.2. Plant Materials for Type-1 RIP Purifications and for Antiviral Assays
4.3. Protein Purification
4.4. Biochemical Analytical Procedures
4.5. Homogeneity of Protein by Capillary Electrophoresis
4.6. Tobacco Necrosis Virus (TNV) Purification
4.7. PD-Ls Treatments and Virus Inoculation
4.8. Evaluation of Antiviral Activity
4.9. Histo-Cytochemistry
4.10. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar]
- Brigotti, M.; Rambelli, F.; Zamboni, M.; Montanaro, L.; Sperti, S. Effect of alpha-sarcin and ribosome-inactivating proteins on the interaction of elongation factors with ribosomes. Biochem. J. 1989, 257, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Polito, L.; Lubelli, C.; Barbieri, L.; Parente, A.; Stirpe, F. Ribosome-inactivating and adenine polynucleotide glycosylase activities in Mirabilis jalapa L. tissues. J. Biol. Chem. 2002, 277, 13709–13716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Ragucci, S.; Russo, R.; Di Maro, A.; Ferreras, J.M. Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- De Zaeytijd, J.; Van Damme, E.J. Extensive evolution of cereal ribosome-inactivating proteins translates into unique structural features, activation mechanisms, and physiological roles. Toxins 2017, 9, 123. [Google Scholar] [CrossRef]
- Peumans, W.J.; Shang, C.; Van Damme, E.J.M. Updated model of the molecular evolution of RIP genes. In Ribosome-Inactivating Proteins; Stirpe, F., Lappi, D.A., Eds.; John Wiley & Sons, Inc.: Ames, IA, USA, 2014; pp. 134–150. [Google Scholar]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-inactivating proteins: An overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar]
- Ferreras, J.M.; Citores, L.; Iglesias, R.; Jiménez, P.; Girbés, T. Sambucus ribosome-inactivating proteins and lectins. In Toxic Plant Proteins; Lord, J.M., Hartley, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 107–131. [Google Scholar]
- Hey, T.D.; Hartley, M.; Walsh, T.A. Maize ribosome-inactivating protein (b-32). Homologs in related species, effects on maize ribosomes, and modulation of activity by pro-peptide deletions. Plant Physiol. 1995, 107, 1323–1332. [Google Scholar] [CrossRef] [Green Version]
- Stirpe, F. Ribosome-inactivating proteins. Toxicon 2004, 44, 371–383. [Google Scholar] [CrossRef]
- Zeng, M.; Zheng, M.; Lu, D.; Wang, J.; Jiang, W.; Sha, O. Anti-tumor activities and apoptotic mechanism of ribosome-inactivating proteins. Chin. J. Cancer 2015, 34, 325–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vago, R.; Marsden, C.J.; Lord, J.M.; Ippoliti, R.; Flavell, D.J.; Flavell, S.-U.; Ceriotti, A.; Fabbrini, M.S. Saporin and ricin A chain follow different intracellular routes to enter the cytosol of intoxicated cells. FEBS J. 2005, 272, 4983–4995. [Google Scholar] [CrossRef] [PubMed]
- de Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rust, A.; Partridge, L.J.; Davletov, B.; Hautbergue, G.M. The use of plant-derived ribosome inactivating proteins in immunotoxin development: Past, present and future generations. Toxins 2017, 9, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polito, L.; Djemil, A.; Bortolotti, M. Plant toxin-based immunotoxins for cancer therapy: A short overview. Biomedicines 2016, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Zhou, Y.-K.; Ji, Z.-L.; Chen, X.-R. The plant ribosome-inactivating proteins play important roles in defense against pathogens and insect pest attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Fabbrini, M.S.; Katayama, M.; Nakase, I.; Vago, R. Plant ribosome-inactivating proteins: Progesses, challenges and biotechnological applications (and a few digressions). Toxins 2017, 9, 314. [Google Scholar] [CrossRef] [Green Version]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef]
- Lapadula, W.J.; Ayub, M.J. Ribosome inactivating proteins from an evolutionary perspective. Toxicon 2017, 136, 6–14. [Google Scholar] [CrossRef]
- Parente, A.; Chambery, A.; Di Maro, A.; Russo, R.; Severino, V. Ribosome-inactivating proteins from Phytolaccaceae. In Ribosome-Inactivating Proteins; Stirpe, F., Lappi, D.A., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 28–43. [Google Scholar]
- Domashevskiy, A.V.; Goss, D.J. Pokeweed antiviral protein, a ribosome inactivating protein: Activity, inhibition and prospects. Toxins 2015, 7, 274–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, P.E.; Bonness, M.S.; Lu, H.; Mabry, T.J. Protoplasts from Phytolacca dodecandra L’Herit (endod) and P. americana L. (pokeweed). Plant Cell Rep. 1996, 15, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Corrado, G.; Bovi, P.D.; Ciliento, R.; Gaudio, L.; Di Maro, A.; Aceto, S.; Lorito, M.; Rao, R. Inducible expression of a Phytolacca heterotepala ribosome-inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology 2005, 95, 206–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Maro, A.; Chambery, A.; Daniele, A.; Casoria, P.; Parente, A. Isolation and characterization of heterotepalins, type 1 ribosome-inactivating proteins from Phytolacca heterotepala leaves. Phytochemistry 2007, 68, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.H.; Song, S.K.; Choi, K.W.; Lee, J.S. Expression of a cDNA encoding Phytolacca insularis antiviral protein confers virus resistance on transgenic potato plants. Mol. Cells 1997, 7, 807–815. [Google Scholar] [PubMed]
- Parente, A.; Berisio, R.; Chambery, A.; Di Maro, A. Type 1 ribosome-Inactivating Proteins from the ombú tree (Phytolacca dioica L.). In Toxic Plant Proteins; Lord, J.M., Hartley, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 79–106. [Google Scholar]
- Chambery, A.; Di Maro, A.; Parente, A. Primary structure and glycan moiety characterization of PD-Ss, type 1 ribosome-inactivating proteins from Phytolacca dioica L. seeds, by precursor ion discovery on a Q-TOF mass spectrometer. Phytochemistry 2008, 69, 1973–1982. [Google Scholar] [CrossRef]
- Di Maro, A.; Berisio, R.; Ruggiero, A.; Tamburino, R.; Severino, V.; Zacchia, E.; Parente, A. Structural and enzymatic properties of an in vivo proteolytic form of PD-S2, type 1 ribosome-inactivating protein from seeds of Phytolacca dioica L. Biochem. Biophys. Res. Commun. 2012, 421, 514–520. [Google Scholar] [CrossRef]
- Di Maro, A.; Valbonesi, P.; Bolognesi, A.; Stirpe, F.; De Luca, P.; Siniscalco Gigliano, G.; Gaudio, L.; Delli Bovi, P.; Ferranti, P.; Malorni, A.; et al. Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 1999, 208, 125–131. [Google Scholar] [CrossRef]
- Di Maro, A.; Chambery, A.; Carafa, V.; Costantini, S.; Colonna, G.; Parente, A. Structural characterization and comparative modeling of PD-Ls 1-3, type 1 ribosome-inactivating proteins from summer leaves of Phytolacca dioica L. Biochimie 2009, 91, 352–363. [Google Scholar] [CrossRef]
- Parente, A.; Conforto, B.; Di Maro, A.; Chambery, A.; De Luca, P.; Bolognesi, A.; Iriti, M.; Faoro, F. Type 1 ribosome-inactivating proteins from Phytolacca dioica L. leaves: Differential seasonal and age expression, and cellular localization. Planta 2008, 228, 963–975. [Google Scholar] [CrossRef]
- Russo, R.; Chambery, A.; Severino, V.; Parente, A.; Di Maro, A. Structural characterization of dioicin 1 from Phytolacca dioica L. gains novel insights into phylogenetic relationships of Phytolaccaceae type 1 RIPs. Biochem. Biophys. Res. Commun. 2015, 463, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Faoro, F.; Conforto, B.; Di Maro, A.; Parente, A.; Iriti, M. Activation of plant defence response contributes to the antiviral activity of Diocin 2 from Phytolacca dioica. In Proceedings of the IOBC/WPRS Working Group “Induced Resistance in Plants Against Insects and Diseases”, Crete, Greece, 27–29 April 2006; Volume 44, pp. 53–57. [Google Scholar]
- Pizzo, E.; Zanfardino, A.; Di Giuseppe, A.M.A.; Bosso, A.; Landi, N.; Ragucci, S.; Varcamonti, M.; Notomista, E.; Di Maro, A. A new active antimicrobial peptide from PD-L4, a type 1 ribosome inactivating protein of Phytolacca dioica L.: A new function of RIPs for plant defence? FEBS Lett. 2015, 589, 2812–2818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzo, E.; Pane, K.; Bosso, A.; Landi, N.; Ragucci, S.; Russo, R.; Gaglione, R.; Torres, M.D.T.; de la Fuente-Nunez, C.; Arciello, A.; et al. Novel bioactive peptides from PD-L1/2, a type 1 ribosome inactivating protein from Phytolacca dioica L. Evaluation of their antimicrobial properties and anti-biofilm activities. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Rubino, L.; Matelli, G.P. Necrovirus. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: Cambridge, MA, USA, 2010; pp. 233–235. [Google Scholar]
- Iriti, M.; Faoro, F. Abscisic acid is involved in chitosan-induced resistance to tobacco necrosis virus (TNV). Plant Physiol. Biochem. 2008, 46, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- Iriti, M.; Sironi, M.; Gomarasca, S.; Casazza, A.P.; Soave, C.; Faoro, F. Cell death-mediated antiviral effect of chitosan in tobacco. Plant Physiol. Biochem. 2006, 44, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Battelli, M.G.; Stirpe, F. Ribosome-inactivating poteins from plants. In Antiviral Proteins in Higher Plants; Chessin, M., DeBorde, D., Zipf, A., Eds.; CRC Press: Boca Raton, FL, USA, 1995; pp. 39–64. [Google Scholar]
- Stirpe, F.; Barbieri, L.; Gorini, P.; Valbonesi, P.; Bolognesi, A.; Polito, L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996, 382, 309–312. [Google Scholar] [CrossRef] [Green Version]
- Picard, D.; Kao, C.C.; Hudak, K.A. Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 2005, 280, 20069–20075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landi, N.; Pacifico, S.; Ragucci, S.; Iglesias, R.; Piccolella, S.; Amici, A.; Di Giuseppe, A.M.A.; Di Maro, A. Purification, characterization and cytotoxicity assessment of Ageritin: The first ribotoxin from the basidiomycete mushroom Agrocybe aegerita. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1113–1121. [Google Scholar] [CrossRef]
- Zhang, L.; French, R.; Langenberg, W.G. Molecular cloning and sequencing of the coat protein gene of a Nebraskan isolate of tobacco necrosis virus: The deduced coat protein sequence has only moderate homology with those of strain A and strain D. Arch. Virol. 1993, 132, 291–305. [Google Scholar] [CrossRef]
- Faoro, F.; Iriti, M. Cell death behind invisible symptoms: Early diagnosis of ozone injury. Biol. Plant. 2005, 49, 585–592. [Google Scholar] [CrossRef]






| Inoculation Method | % Reduction of TNV-Lesions |
|---|---|
| TNV + PD-L1 2 µg/mL on the same leaf surface | 89.1 ± 3 |
| TNV + PD-L1 10 µg/mL on the same leaf surface | 95.8 ± 2 |
| TNV + PD-L4 2 µg/mL on the same leaf surface | 98.7 ± 4 |
| TNV + PD-L4 10 µg/mL on the same leaf surface | 99.7 ± 2 |
| TNV on the adaxial leaf surface and PD-L4 on the abaxial | 72.6 ± 3 |
| TNV on the abaxial leaf surface and PD-L4 on the adaxial | 78.0 ± 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulgari, D.; Landi, N.; Ragucci, S.; Faoro, F.; Di Maro, A. Antiviral Activity of PD-L1 and PD-L4, Type 1 Ribosome Inactivating Proteins from Leaves of Phytolacca dioica L. in the Pathosystem Phaseolus vulgaris–Tobacco Necrosis Virus (TNV). Toxins 2020, 12, 524. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080524
Bulgari D, Landi N, Ragucci S, Faoro F, Di Maro A. Antiviral Activity of PD-L1 and PD-L4, Type 1 Ribosome Inactivating Proteins from Leaves of Phytolacca dioica L. in the Pathosystem Phaseolus vulgaris–Tobacco Necrosis Virus (TNV). Toxins. 2020; 12(8):524. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080524
Chicago/Turabian StyleBulgari, Daniela, Nicola Landi, Sara Ragucci, Franco Faoro, and Antimo Di Maro. 2020. "Antiviral Activity of PD-L1 and PD-L4, Type 1 Ribosome Inactivating Proteins from Leaves of Phytolacca dioica L. in the Pathosystem Phaseolus vulgaris–Tobacco Necrosis Virus (TNV)" Toxins 12, no. 8: 524. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080524