Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus scutulatus × viridis Hybrid Zone in Southwestern New Mexico
Abstract
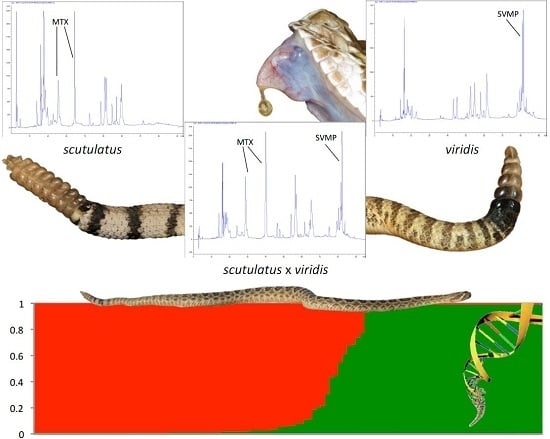
:1. Introduction
2. Results
2.1. Morphology
2.2. Molecular Evidence of Hybridization
2.3. Detection of Mojave Toxin
3. Discussion
4. Materials and Methods
4.1. Morphological Methods
4.2. Molecular Analysis of Hybridization
4.3. Determination of Mojave Toxin Presence
4.4. Determination of Mojave Toxin in the Venom
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MTX | Mojave toxin |
| PLA2 | Phospholipase A2 |
| RP-HPLC | Reverse-phase high performance liquid chromatography |
| SVMP | Snake venom metalloproteinases |
Appendix A
| Locus | Forward | Reverse |
|---|---|---|
| ND4 | ND4: CACCTATGACTACCAAAAGCTCATGTAGAAGC | Leu: CATTACTTTTACTTGGATTTGCACCA |
| H12763V: TTCTATCACTTGGATTTGCACCA | ||
| NT3 | NTF3_SC_F: CGAGGTTTTGCACTGGGAAT | NTF3_SC_R: GCATTTCTGTGTGGCATCCA |
| R35 | R35_F: GACTGTGGAYGAYCTGATCAGTGTGGTGCC | R35_R: GCCAAAATGAGSGAGAARCGCTTCTGAGC |
| SELT | SELT_F: GTTATYAGCCAGCGGTACCCAGACATCCG | SELT_R: GCCTATTAAYACTAGTTTGAAGACTGACAG |
| ETS | ETS_F: CCATCAACAGACACACAGG | ETS_R: GTCTGCTTTTTACTTTGCG |
| MTXa | MTXa2_F: TGCGGGGAGAAGTGGTATTT | MTXa4_R: GCAATTTTCGGGCGAGAACC |
| MTXb | MTXb2_F: ACCTGCTGCAATTCAACAAGA | MTXb4_R: CGAGAGTCCGGGTAAAACAT |
| PCR Parameter | ND4 | NT3/R35 | SELT/ETS | MTXa/MTXb |
|---|---|---|---|---|
| 1. Initial denaturation | 94°-2 m | 94°-2 m | 94°-2 m | 94°-2 m |
| 2. Denaturation | 94°-30 s | 94°-30 s | 94°-30 s | 94°-30 s |
| 3. Annealing | 57°-30 s | 55°-1 m | 47°-1 m | 59°-30 s |
| 4. Extension | 72°-1 m | 72°-1 m | 72°-1 m | 72°-1.5 m |
| 5. No. Cycles (2–4) | 40 | 35 | 35 | 35 |
| 6. Final extension | 72°-5 m | 72°-5 m | 72°-5 m | 72°-5 m |
References
- Chippaux, J.P.; Williams, V.; White, J. Snake Venom Variability: Methods of study, results and interpretation. Toxicon: Off. J. Int. Soc. Toxinol. 1991, 29, 1279–1303. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Warrell, D.A. Snake venoms in science and clinical medicine. 1. Russell’s viper: Biology, venom and treatment of bites. Trans. R. Soc. Trop. Med. Hyg. 1989, 83, 732–740. [Google Scholar] [CrossRef]
- Warrell, D.A. Geographical and intraspecies variation in the clinical manifestations of envenoming by snakes. Symp. Zool. Soc. Lond. 1997, 70, 189–203. [Google Scholar]
- Massey, D.J.; Calvete, J.J.; Sánchez, E.E.; Sanz, L.; Richards, K.; Curtis, R.; Boesen, K. Venom variability and envenoming severity outcomes of the Crotalus. scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteom. 2012, 75, 2576–2587. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Nakashima, K.; Nobuhisa, I.; Deshimaru, M.; Shimohigashi, Y.; Fukumaki, Y.; Sakaki, Y.; Hattori, S.; Ohno, M. Accelerated evolution of snake venom phospholipase A2 isozymes for acquisition of diverse physiological functions. Toxicon: Off. J. Int. Soc. Toxinol. 1996, 34, 1229–1236. [Google Scholar] [CrossRef]
- Lynch, V.J. Inventing an arsenal: Adaptive evolution and neofunctionalization of snake venom phospholipase A(2) genes. BMC Evol. Biol. 2007, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Rossiter, W. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus. rattlesnakes with divergent diets. J. Mol. Evol. 2008, 66, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, R.S.; Pook, C.E.; Malhotra, A. Phylogeography of the Russell’s viper (Daboia. russelii) complex in relation to variation in the colour pattern and symptoms of envenoming. Herpetol. J. 2007, 17, 209–218. [Google Scholar]
- Jorge da Silva, N.; Aird, S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. C 2001, 128, 425–456. [Google Scholar] [CrossRef]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Co-evolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. B 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Mackessy, S.P. Functional basis of a molecular adaptation: Prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon: Off. J. Int. Soc. Toxinol. 2009, 53, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.L.; Straight, R.C. Mojave rattlesnake Crotalus. scutulatus scutulatus venom: Variation in toxicity with geographical origin. Toxicon: Off. J. Int. Soc. Toxinol. 1978, 16, 81–84. [Google Scholar] [CrossRef]
- McCue, M.D. Enzyme activities and biological functions of snake venoms. Appl. Herpetol. 2005, 2, 109–123. [Google Scholar] [CrossRef]
- Mackessy, S.P. Venom composition in rattlesnakes: Trends and biological significance. In The Biology of the Rattlesnakes; Hayes, W.K., Beaman, K.R., Cardwell, M.D., Bush, S.P., Eds.; Loma Linda University Press: Loma Linda, CA, USA, 2008; pp. 495–510. [Google Scholar]
- Mackessy, S.P.; Williams, K.; Ashton, K.G. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: A case of venom paedomorphosis? Copeia 2003, 2003, 769–782. [Google Scholar] [CrossRef]
- Mackessy, S.P. Evolutionary trends in venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): Toxicity vs. tenderizers. Toxicon: Off. J. Int. Soc. Toxinol. 2010, 55, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.L.; Straight, R.C. Distribution of proteins immunologically similar to Mojave toxin among species of Crotalus and Sistrurus. Toxicon: Off. J. Int. Soc. Toxinol. 1985, 23, 28. [Google Scholar]
- Glenn, J.L.; Straight, R.C. Intergradation of two different venom populations of the Mojave rattlesnake (Crotalus scutulatus scutulatus) in Arizona. Toxicon: Off. J. Int. Soc. Toxinol. 1989, 27, 411–418. [Google Scholar] [CrossRef]
- Glenn, J.L.; Straight, R.C.; Wolfe, M.C.; Hardy, D.L. Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon: Off. J. Int. Soc. Toxinol. 1983, 21, 119–130. [Google Scholar] [CrossRef]
- Powell, R.L.; Lieb, C.S.; Rael, E.D. Geographic distribution of Mojave toxin and Mojave toxin subunits among selected Crotalus species. Venom composition in rattlesnakes: Trends and biological significance. In The Biology of the Rattlesnakes; Hayes, W.K., Beaman, K.R., Cardwell, M.D., Bush, S.P., Eds.; Loma Linda University Press: Loma Linda, CA, USA, 2008; pp. 537–550. [Google Scholar]
- Durban, J.; Pérez, A.; Sanz, L.; Gómez, A.; Bonilla, F.; Rodríguez, S.; Chacón, D.; Sasa, M.; Angulo, Y.; Gutiérrez, J.M.; et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake Crotalus simus simus. BMC Genom. 2013, 14, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunagar, K.; Undheim, E.A.B.; Scheib, H.; Gren, E.C.K.; Cochran, C.; Person, C.E.; Koludarov, I.; Kelln, W.; Hayes, W.K.; King, G.F.; et al. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): Biodiscovery, clinical and evolutionary implications. J. Proteom. 2014, 99, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Saravia, P.; Rojas, E.; Arce, V.; Guevara, C.; López, J.C.; Chaves, E.; Velásquez, R.; Rojas, G.; Gutiérrez, J.M. Geographic and ontogenic variability in the venom of the neotropical rattlesnake Crotalus durissus: pathophysiological and therapeutic implications. Rev. Biol. Trop. 2002, 50, 337–346. [Google Scholar] [PubMed]
- Calvete, J.J.; Sanz, L.; Cid, P.; de la Torre, P.; Flores-Díaz, M.; dos Santos, M.C.; Borges, A.; Bremo, A.; Angulo, Y.; Lomonte, B.; Alape-Girón, A.; Gutiérrez, J.M. Snake venomics of the Central American Rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010, 9, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.L.; Straight, R.C.; Wolt, T.B. Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comp. Biochem. Physiol. C 1994, 107, 337–346. [Google Scholar] [CrossRef]
- Wooldridge, B.J.; Pineda, G.; Banuelas-Ornelas, J.J.; Dagda, R.K.; Gasanov, S.E.; Rael, E.D.; Lieb, C.S. Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comp. Biochem. Physiol. B 2001, 130, 169–179. [Google Scholar] [CrossRef]
- Sánchez, E.E.; Galán, J.A.; Powell, R.L.; Reyes, S.R.; Soto, J.G.; Russell, W.K.; Russell, D.H.; Pérez, J.C. Disintegrin, hemorrhagic, and proteolytic activities of Mohave rattlesnake, Crotalus scutulatus scutulatus venoms lacking Mojave toxin. Comp. Biochem. Physiol. C 2005, 141, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Wray, K.P.; McGivern, J.J.; Margres, M.J. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon: Off. J. Int. Soc. Toxinol. 2015, 98, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Lewontin, R.C.; Birch, L.C. Hybridization as a source of variation for adaptation to new environments. Evolution 1966, 20, 315–336. [Google Scholar] [CrossRef]
- Hedrick, P.W. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 2013, 22, 4606–4618. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H. Gene flow past a cline. Heredity 1979, 43, 333–339. [Google Scholar] [CrossRef]
- Harrison, R.G.; Larson, E.L. Hybridisation, introgression, and the nature of species boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Uecker, H.; Setter, D.; Hermisson, J. Adaptive gene introgression after secondary contact. J. Math. Biol. 2015, 70, 1523–1580. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Ann. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar] [CrossRef]
- Glenn, J.L.; Straight, R.C. Venom characteristics as an indicator of hybridisation between Crotalus viridis viridis and Crotalus scutulatus scutulatus in New Mexico. Toxicon: Off. J. Int. Soc. Toxinol. 1990, 28, 857–862. [Google Scholar] [CrossRef]
- Aird, S.D.; Thirkhill, L.J.; Seebart, C.S.; Kaiser, I.I. Venoms and Morphology of Western Diamondback/Mojave Rattlesnake Hybrids. J. Herpetol. 1989, 23, 131–141. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Parmelee, J.; Guo, Y.-W.; Tsai, I.-H. Absence of phospholipase A2 in most Crotalus horridus venom due to translation blockage: Comparison with Crotalus horridus atricaudatus venom. Toxicon: Off. J. Int. Soc. Toxinol. 2010, 56, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, I.; Bouchier, C.; Garrigues, T.; Wisner, A.; Choumet, V. Sequences and structural organization of phospholipase A2 genes from Vipera aspis aspis, V. aspis zinnikeri and Vipera berus berus venom. Identification of the origin of a new viper population based on ammodytin I1 heterogeneity. Eur. J. Biochem. 2003, 270, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Ferquel, E.; de Haro, L.; Jan, V.; Guillemin, I.; Jourdain, S.; Teynié, A.; d’Alayer, J.; Choumet, V. Reappraisal of Vipera aspis venom neurotoxicity. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Grenard, S. Is rattlesnake venom evolving? Nat. Hist. Mag. 2000, 109, 44–46. [Google Scholar]
- Hayes, W.K.; Mackessy, S.P. Sensationalistic journalism and tales of snakebite: Are rattlesnakes rapidly evolving more toxic venom? Wild. Environ. Med. 2010, 21, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.A.; Sánchez, E.E.; Rodríguez-Acosta, A.; Pérez, J.C. Neutralization of venoms from two Southern Pacific Rattlesnakes (Crotalus helleri) with commercial antivenoms and endothermic animal sera. Toxicon: Off. J. Int. Soc. Toxinol. 2004, 43, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Klauber, L.M. Rattlesnakes: Their Habits, Life Histories, and Influence on Mankind, 2nd ed.; University of California Press: Berkeley, CA, USA, 1972. [Google Scholar]
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of Latin America; Comstock: Ithaca, NY, USA/London, UK, 1989. [Google Scholar]
- Campbell, J.A.; Brodie, E.D.; Barker, D.G.; Price, A.H. An apparent natural hybrid rattlesnake and Crotalus willardi (Viperidae) from the Peloncillo Mountains of southwestern New Mexico. Herpetologica 1989, 45, 344–349. [Google Scholar]
- Meik, J.M.; Fontenot, B.E.; Franklin, C.J.; King, C. Apparent natural hybridization between the rattlesnakes Crotalus atrox and C. horridus. Southwest. Nat. 2008, 53, 196–200. [Google Scholar] [CrossRef]
- Montgomery, W.B.; Schuett, G.W.; Douglas, M.R.; Douglas, M.E. Crotalus atrox x Crotalus horridus (Western diamond-backed rattlesnake x timber rattlesnake). Natural Hybrid. Herpetol. Rev. 2013, 44, 689. [Google Scholar]
- Murphy, R.W.; Crabtree, B.C. Genetic Identification of a Natural Hybrid Rattlesnake: Crotalus scutulatus scutulatus x C. viridis viridis. Herpetologica 1988, 44, 119–123. [Google Scholar]
- Jacob, J.S. An evaluation of the possibility of hybridization between the rattlesnakes Crotalus atrox and C. scutulatus in the Southwestern United States. Southwest. Nat. 1977, 22, 469–485. [Google Scholar] [CrossRef]
- Degenhardt, W.G.; Painter, C.W.; Price, A.H. Amphibians and Reptiles of New Mexico; University of New Mexico Press: Albuquerque, NM, USA, 2005. [Google Scholar]
- John, T.R.; Smith, L.A.; Kaiser, I.I. Genomic sequences encoding the acidic and basic subunits of Mojave toxin: Unusually high sequence identity of non-coding regions. Gene 1994, 139, 229–234. [Google Scholar] [CrossRef]
- Wilkinson, J.A.; Glenn, J.L.; Straight, R.C.; Sites, J.W. Distribution and genetic variation in venom A and B populations of the Mojave rattlesnake (Crotalus scutulatus scutulatus) in Arizona. Herpetologica 1991, 47, 54–68. [Google Scholar]
- Tarroso, P.; Pereira, R.J.; Martínez-Freiría, F.; Godinho, R.; Brito, J.C. Hybridization at an ecotone: Ecological and genetic barriers between three Iberian vipers. Mol. Ecol. 2014, 23, 1108–1123. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Wüster, W.; Cook, D.A.N.; Bolton, F.M.S.; King, S.I.; Pla, D.; Sanz, L.; Calvete, J.J.; Harrison, R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. PNAS 2014, 111, 9205–9210. [Google Scholar] [CrossRef] [PubMed]
- Minton, S.A.; Weinstein, S.A. Geographic and ontogenetic variation in venom of the western diamondback rattlesnake (Crotalus atrox). Toxicon. 1986, 24, 71–80. [Google Scholar] [CrossRef]
- Vidal, N.; Delmas, A.-S.; David, P.; Cruaud, C.; Couloux, A.; Hedges, S.B. The phylogeny and classification of caenophidian snakes inferred from seven nuclear protein-coding genes. C. R. Biol. 2007, 330, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Diaz, J. Identification of single copy nuclear DNA markers for North American pit vipers. Mol. Ecol. Resour. 2010, 10, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, J.R. Multilocus species delimitation and species tree inference within the western rattlesnake (Crotalus viridis) species complex. Unpublished MS Thesis, San Diego State University, San Diego, CA, USA, May 2013. [Google Scholar]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar]
- Dmitriev, D.A.; Rakitov, R.A. Decoding of superimposed traces produced by direct sequencing of heterozygous indels. PLoS Comput. Biol. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Human Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Scheet, P. Accounting for Decay of Linkage Disequilibrium in Haplotype Inference and Missing-Data Imputation. Am. J. Human Genet. 2005, 76, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Flot, J.-F. SeqPHASE: A web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol. Ecol. Resour. 2010, 10, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Hubisz, M.; Falush, D.; Stephens, M.; Pritchard, J. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure: Extensions to linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]





| Sequence parameters | ND4 | NT3 | R35 | SELT | ETS |
|---|---|---|---|---|---|
| Length (bp) | 635 | 538 | 531 | 346 | 653 |
| # haplotypes | 57 | 39 | 18 | 20 | 26 |
| # variable positions | 139 | 31 | 15 | 17 | 29 |
| 1. Number of internasals contacting rostral scale |
| 2. Minimum number of scales separating posteriormost canthals |
| 3. Minimum number of scales separating supraoculars |
| 4. Number of scales contacting the inner edge of the supraoculars |
| 5. Number of dark (defined as noticeably darker than body markings) bands on tail |
| 6. Number of light (defined as noticeably lighter than body ground colour) bands on tail |
| 7. Maximum width in dorsal scale lengths along a single scale row (excluding the vertebral row) of the posteriormost black band not contacting the rattle fringe. |
| 8. Maximum width in dorsal scale lengths along a single scale row (excluding the vertebral row) of the light band anterior to 7. |
| 9. Basal rattle segment entirely light (0), black (1) or partly light, partly black (0.5). |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zancolli, G.; Baker, T.G.; Barlow, A.; Bradley, R.K.; Calvete, J.J.; Carter, K.C.; De Jager, K.; Owens, J.B.; Price, J.F.; Sanz, L.; et al. Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus scutulatus × viridis Hybrid Zone in Southwestern New Mexico. Toxins 2016, 8, 188. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8060188
Zancolli G, Baker TG, Barlow A, Bradley RK, Calvete JJ, Carter KC, De Jager K, Owens JB, Price JF, Sanz L, et al. Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus scutulatus × viridis Hybrid Zone in Southwestern New Mexico. Toxins. 2016; 8(6):188. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8060188
Chicago/Turabian StyleZancolli, Giulia, Timothy G. Baker, Axel Barlow, Rebecca K. Bradley, Juan J. Calvete, Kimberley C. Carter, Kaylah De Jager, John Benjamin Owens, Jenny Forrester Price, Libia Sanz, and et al. 2016. "Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus scutulatus × viridis Hybrid Zone in Southwestern New Mexico" Toxins 8, no. 6: 188. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8060188