Published online Oct 1, 2004. doi: 10.3748/wjg.v10.i19.2831
Revised: April 9, 2004
Accepted: April 16, 2004
Published online: October 1, 2004
AIM: To investigate the effects of herbal compound 861 (Cpd 861) on cell proliferation in human hepatic stellate cells (LX-2) and human hepatocellular liver carcinoma cells (HepG2), and expression of α-smooth muscle actin (α-SMA) in LX-2 cells.
METHODS: LX-2 and HepG2 cells were incubated with various concentrations of Cpd 861 (0.1-0.003 mg/mL) for 1, 2, 3, 5 and 7 d. Cell proliferation was analyzed by 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. Effects of Cpd861 on the expression of α-SMA mRNA in LX-2 cells were measured by real-time quantitative PCR method using SYBR Green I technology.
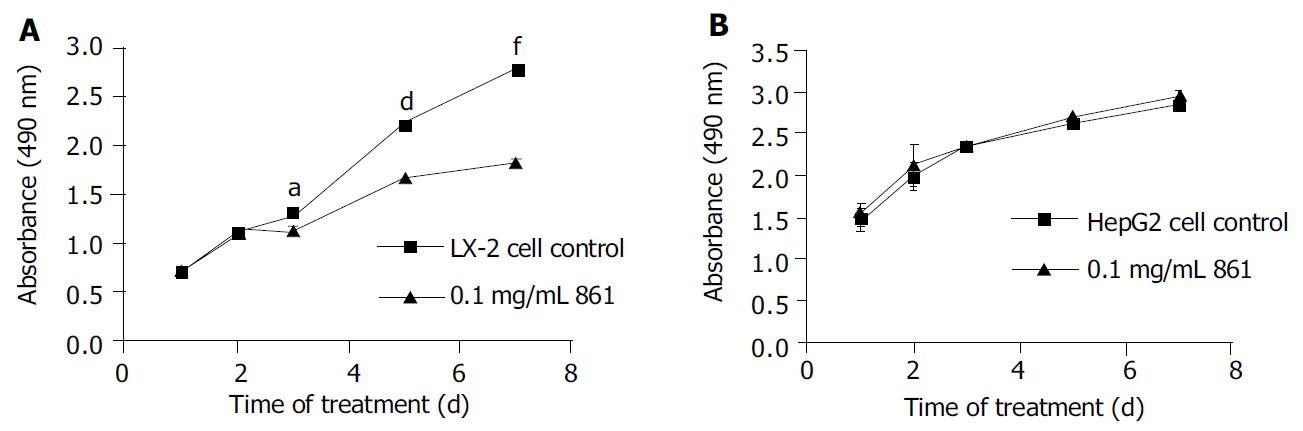
RESULTS: Cpd 861, at 0.1 mg/mL, significantly inhibited LX-2 cell proliferation (15% decrease relative to control, P < 0.05) after 3 d of incubation. The inhibitory effects seemed to increase with the treatment time (25% decrease after 5 d of incubation and 35% decrease after 7 d of incubation, P < 0.01). However, Cpd 861 did not affect HepG2 cell proliferation at the same concentration used for LX-2 cells. The expression levels of α-SMA mRNA decreased significantly when LX-2 cells were exposed to Cpd 861 for 48 h (59% decrease relative to control, P < 0.05) or 72 h (60% decrease relative to control, P < 0.01).
CONCLUSION: Cpd 861 can significantly inhibit LX-2 cell proliferation in a dose-dependant manner, and reduce the expression levels of α-SMA mRNA in LX-2 cells. Since hepatic cell proliferation and high level of α-SMA are associated with liver fibrosis, the results suggest that Cpd 861 may be useful in the treatment of this disease.
- Citation: Wang L, Wang J, Wang BE, Xiao PG, Qiao YJ, Tan XH. Effects of herbal compound 861 on human hepatic stellate cell proliferation and activation. World J Gastroenterol 2004; 10(19): 2831-2835
- URL: https://www.wjgnet.com/1007-9327/full/v10/i19/2831.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i19.2831
Hepatic fibrosis is a reversible wound healing response to chronic liver injury due to a variety of insults, including viral hepatitis (especially hepatitis B and C), alcohol abuse, drugs, metabolic diseases, autoimmune attack of hepatocytes or congenital abnormalities[1,2]. As shown in many recent studies, hepatic stellate cells (previously called Ito cells, lipocytes, perisinusoidal cells, or fat-storing cells) are primary cell types to mediate fibrogenesis[1,2]. In normal liver, hepatic stellate cells (HSCs) are nonparenchymal quiescent cells with functions to store vitamin A. Following liver injury of any etiology, HSCs undergo a process of activation, transform from quiescent vitamin A-rich cells to proliferative, fibrogenic, contractile myofibroblasts. Activated HSCs lose lipid droplets, feature high level expression of α-smooth muscle actin (α-SMA), and also are responsible for the deposition of the majority of excess extracellular matrix (ECM, predominantly types I and III collagen), which leads to form action of scar tissue in the fibrotic liver[1-4].
Herbal compound 861 (Cpd 861) is an extract of 10 herbs with Salvia miltiorrhiza, Astragalus membranaceus and Spatholobus suberectus as its chief components. The recipe of this mixed compound was based on the therapeutically indications of Chinese medicine. Clinical studies showed Cpd 861 could significantly improve clinical symptoms in hepatic fibrosis patients as well as the biochemical parameters associated with diseases in clinical tests for patients[5,6]. Experimental researches also showed that Cpd 861 could inhibit cell proliferation, reduce the level of α-SMA and reverse the process of liver fibrosis on rat model[5-7]. In this study, we aimed to investigate the effects of Cpd 861 on LX-2 and HepG2 cell proliferation and expression of α-SMA in LX-2 cells.
Human activated hepatic stellate cell line LX-2 was a gift from Dr. Friedman of Mount Sinai School of Medicine. LX-2 cells were a low-passaged human cell line from normal human stellate cells that were spontaneously immortalized[8,9]. The cells exhibited typical features of stellate cells and expressed α-SMA under all culture conditions and were regarded as at least partially activated even after immediate replating. LX-2 cells underwent further activation during growth and expansion on plastic surfaces[8].
Human hepatocellular carcinoma cell line HepG2 was purchased from the Chinese Academy of Medical Sciences (Beijing, China). Dulbecco’s modified Eagle’s medium (DMEM), L-glutamine and streptomycin were purchased from Gibco, Invitrogen, Carlsbad, CA. Fetal bovine serum (FBS) was from Hyclone, USA. Penicillin and phenazine methosulfate (PMS) were from Sigma, USA. MTS and oligo (dT)15 primers were acquired from Promega, USA. TRIzol reagent and Moloney murine leukemia virus reverse transcriptase (M-MLV RT) were the products of Invitrogen, CA. SYBR Green I was purchased from OPE Technology Development Company, Shanghai, China. Hotstar Taq DNA polymerase was purchased from TW-Biotech, China.
LX-2 cells were cultured in DMEM supplemented with 50 mL/L heat-inactivated FBS, 200 mmol/L L-glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin. HepG2 cells were cultured in DMEM containing 100 mL/L FBS, 200 mmol/L L-glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin.
MTS assay was used to determine the effect of Cpd 861 on the proliferation of LX-2 and HepG2 cells. In metabolically active cells, MTS was reduced by dehydrogenase enzymes into an aqueous, soluble formazan product. The absorbance was measured directly at 490 nm from 96-well assay plates without additional processing. The quantity of formazan was considered to be directly proportional to the number of viable cells in the culture[10,11].
LX-2 and HepG2 cells were seeded into 96-well tissue culture plates containing 100 µL DMEM containing 50 mL/L FBS (5000 LX-2 cells/well and 8000 HepG2 cells/well). After 24 h of incubation (37 °C, 50 mL/L CO2), the medium was carefully removed and 100 µL fresh medium containing various concentrations of Cpd 861 was added into the wells. The cells were treated continuously with Cpd 861 for 1, 2, 3, 5 and 7 d and the medium containing Cpd 861 was changed every other day. At the end of experiments, 20 µL/well of combined MTS/PMS solution was added. After 3 h of incubation at 37 °C in a humidified incubator, the absorbance was analyzed on a VERSAmax microplate reader at 490 nm. Absorbance values were the mean ± SE of 3 replicates for each treatment. The cells in only controls and compound controls were included.
LX-2 cells were seeded into 60 mm dishes in DMEM containing 50 mL/L FBS. After 3 d of seeding, the medium was carefully removed and fresh medium containing Cpd 861 (0.01 mg/mL) was added. After 24, 48 and 72 h of Cpd 861 treatment, total RNA was extracted from LX-2 cells using TRIzol reagent as the lysis buffer. Complementary DNA (cDNA) was synthesized using oligo (dT)15 primers and M-MLV RT.
After the reverse-transcription reaction, cDNA templates were amplified by quantitative real-time PCR. The reaction was performed using the iCycler iQ real-time PCR detection system (Bio-Rad, USA) with SYBR Green I. Human α-SMA and G3PDH (glyceraldehyde-3-phosphate dehydrogenase) primers were designed using Primer 3 software (Table 1).
| Oligonucleotide | Oligonucleotide primer sequence | Fragment size |
| α-SMA | ||
| sense | ACT GGG ACG ACA TGG AAA AG | 265 bp |
| antisense | TAG ATG GGG ACA TTG TGG GT | |
| G3PDH | ||
| sense | ACC CAG AAG ACT GTG GAT GG | 125 bp |
| antisense | TTC AGC TCA GGG ATG ACC TT |
Each experiment was performed in 25 µL of reaction volume [1.25 µL of 20 × SYBR Green, 1 µL of first strand cDNA (50 ng RNA), 2 µL of each 5 nmolar primer, 0.75 U Hotstar Taq DNA polymerase, 2.5 µL of 10 × amplification buffer (Mg++ free), 2.5 µL of 25 mmol/L MgCl 2, 0.5 µL of 10 mmol/L solution of four dNTP and 15.1 µL of dH 2O]. In the last tube, 1 µL of ddH 2O was added as a non-template control. The conditions of amplification cycles were as follows: 40 cycles consisting of denaturation at 95 °C for 40 s, annealing at 59 °C (for α-SMA) or at 60 °C (for G3PDH) for 40 s, and extension at 72 °C for 30 s.
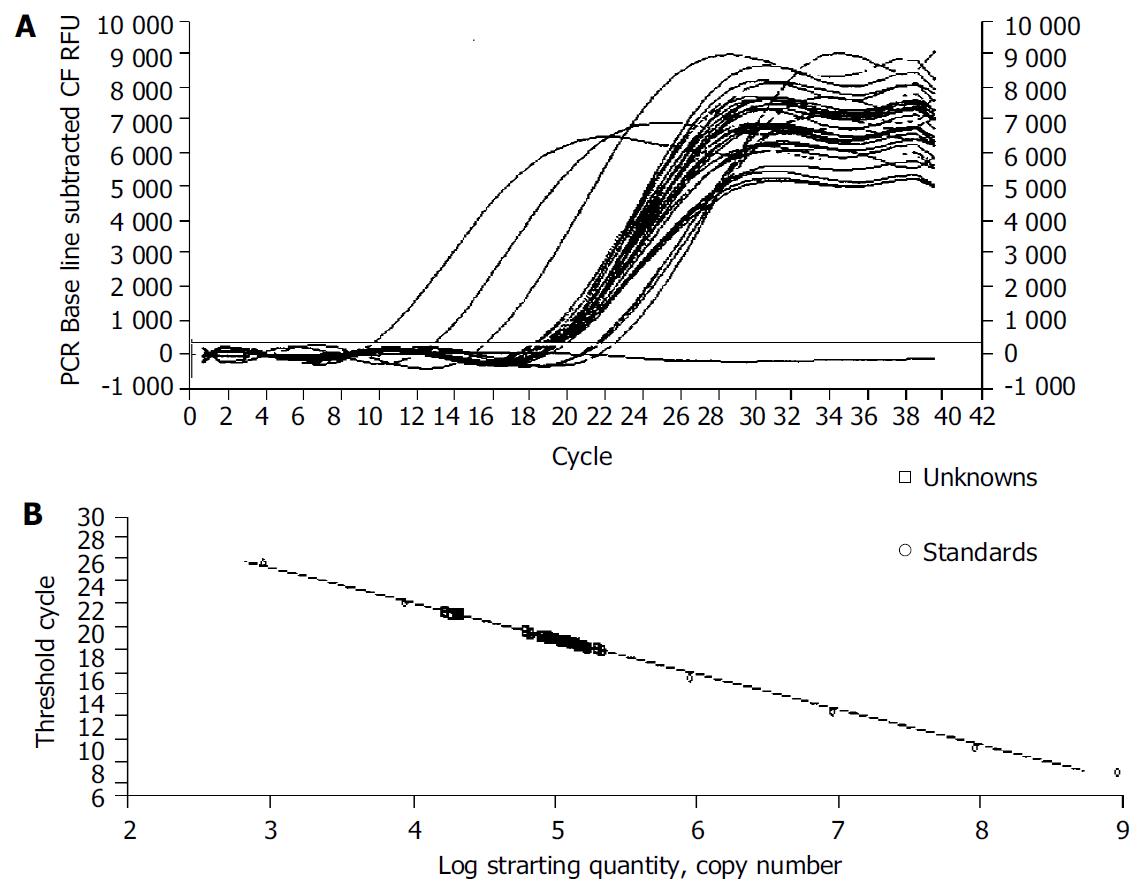
The iCycler apparatus was used to measure the fluorescence of each sample in every cycle at the end of the extension[12]. A series of consecutive 10 fold dilution of α-SMA and G3PDH plasmid DNA ranging from 109 copies/µL to 103 copies/µL were used as the templates for the standard curves. The iCycler software was used to construct the calibration curve by plotting the Ct (threshold cycle) vs the logarithm of the number of copies for each calibrator. The number of copies in unknown samples was calculated by comparing their Cts with the standard calibration curve. The quality and quantities of samples were normalized based on that of G3PDH.
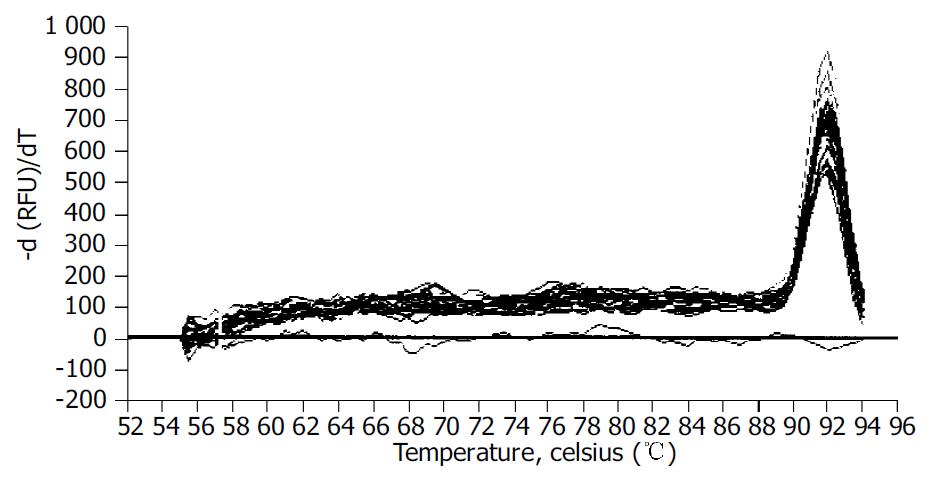
After PCR, a melting curve was obtained by increasing the temperature from 55 °C to 95 °C with a temperature transition rate of 0.1 °C/s. The melting curves of all final PCR products were analyzed. The differences in melting temperature of PCR products allowed us to distinguish genuine products from nonspecific products, and primer dimers. To ensure that the correct product was amplified in the reaction, all samples were also separated on 20 g/L agarose gel electrophoresis. All PCR conditions and primers were optimized to produce a single product of the correct basepair size.
Data were expressed as mean ± SE. Statistical analysis was performed using GraphPad Prism (version 3.0) software. The t test was used for comparison between the groups. P < 0.05 was considered statistically significant.
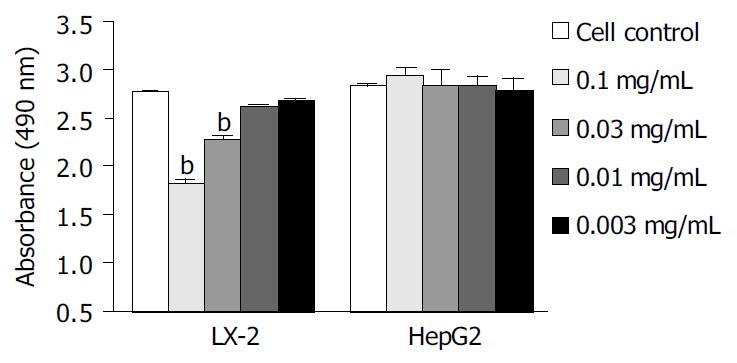
We examined the effects of Cpd 861 on the proliferation of LX-2 and HepG2 cells (Figure 1, Figure 2). Cells were incubated with 0.1 mg/mL, 0.03 mg/mL, 0.01 mg/mL and 0.003 mg/mL of Cpd 861 for different days (Figure 1). Cell proliferation was performed by MTS assay. As shown in Figure 2, the effects of Cpd 861 on LX-2 cell proliferation seemed to be correlated with the dose. A significant proliferation inhibition was observed when Cpd 861 concentration was over 0.03 mg/mL (Figure 2). As shown in Figure 1A, 0.1 mg/mL Cpd 861 significantly inhibited LX-2 cell proliferation (15% decrease relative to control, P < 0.05) after 3 d of incubation. The inhibition effects seemed to increase with the treatment time (25% decrease after 5 d of incubation and 35% decrease after 7 d of incubation, P < 0.01). The inhibition of cell proliferation Cpd 861 at 0.03 mg/mL concentration was also observed on d 3 (15% decrease relative to control) and on d 5 (21% decrease relative to control) (data not shown), and still could be observed on d 7 (18% decrease relative to control) (Figure 2). However, Cpd 861, at 0.01 mg/mL and 0.003 mg/mL concentrations, did not inhibit LX-2 proliferation evidently. The effects of Cpd 861 on LX-2 cell proliferation seemed low at early time points (on day 1 and day 2). We also examined the effects of Cpd 861 on the proliferation of HepG2 cells. As shown in Figure 1B and Figure 2, Cpd 861 did not affect HepG2 cell proliferation.
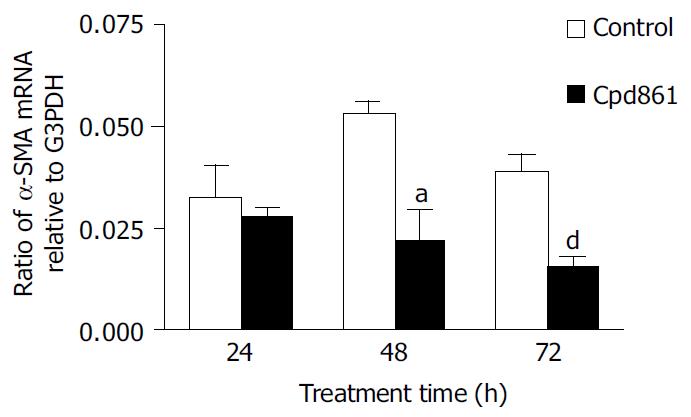
We examined the effect of Cpd 861 on the expression of α-SMA, a phenotypic marker of activated HSCs. The PCR amp/cycle graph and standard curve graph of α-SMA are shown in Figure 3. The standard curve showed a correlation coefficient > 0.99, indicating a precise log-linear relationship. As shown in melt curve graph (Figure 4), single and sharply defined melting curves with narrow peaks were obtained for PCR products of α-SMA gene. Bands visible after electrophoresis on 2% agarose gel and ethidium bromide staining correlated well with the quantitative PCR results. As shown in Figure 5, Cpd 861 could reduce the expression levels of α-SMA in LX-2 cells after treatment. When LX-2 cells were exposed to Cpd 861 for 48 or 72 h, the expression levels of α-SMA decreased significantly (59% decrease relative to control at 48 h, P < 0.05, and 60% decrease relative to control at 72 h, P < 0.01) (Figure 5).
In the liver, activated stellate cells are the key mediators of fibrosis[4]. During hepatic fibrogenesis, HSCs undergo an activation process, becoming highly proliferative and α-SMA positive myofibroblast-like cells.
Cpd 861 is an effective herbal compound for treatment of patients with hepatic fibrosis[5]. Its function to effectively reverse hepatic fibrosis has been confirmed by liver biopsies[5-7]. In other studies, data showed that Cpd 861 also could suppress inflammation and fibrogenesis of the liver tissue[7,13,14] and reduce expression of collagens and fibrosis related cytokines[14-19]. It could also inhibit rat hepatic stellate cell proliferation in a dose-dependent manner and decrease activation of rat HSCs in vivo and in vitro[20,21].
Our results suggested that Cpd 861 could significantly inhibit human hepatic stellate cell LX-2 proliferation in a dose-dependant manner when its concentration was over 0.03 mg/mL. The inhibition function increased obviously with the incubation time. However, the results showed that Cpd 861 did not inhibit HepG2 cell proliferation at the same concentration, indicating that Cpd 861 probably could specifically inhibit hepatic stellate cell proliferation, although further study is needed.
RT-PCR is a technique that has been wildly used to quantify physiological changes in gene expression[22]. However, there is lack of accurate quantification in most of the cases[22,23]. Real-time PCR techniques developed during the last decade could offer much more accurate and precise quantitation of DNA and RNA[23,24]. In our study a reliable and accurate real-time PCR method using SYBR Green I technology[25,26] was taken to measure the effects of Cpd 861 on expression of α-SMA mRNA in LX-2 cells. To quantify the number of molecules of α-SMA mRNA, standard curves were constructed with plasmids containing the fragments for α-SMA and G3PDH as the templates. A high linearity was observed over a dynamic range of at least 5 orders of magnitude (from 109 to 103 copies). All PCR conditions and primers were optimized to produce the target products of the correct basepair size just as shown in melt curves. Quantities of α-SMA were expressed as relative ratio to the copy number of G3PDH. The results indicated that the expression levels of α-SMA mRNA decreased significantly when LX-2 cells were exposed to Cpd 861 at 0.01 mg/mL concentration for 48 or 72 h. Although at the same concentration, Cpd 861 did not inhibit LX-2 cell proliferation.
In conclusion, Cpd 861 can significantly inhibit LX-2 proliferation in a dose-dependant manner when its concentration is over 0.03 mg/mL. Additionally, Cpd 861 can reduce the expression levels of α-SMA mRNA in LX-2 cells and this function seems independent of its effects on inhibition of cell proliferation.
We thank Dr. Friedman and Mount Sinai, School of Medicine, for their help in providing the LX-2 cell line.
Co-correspondents: Jian Wang
Edited by Kumar M and Wang XL Proofread by Xu FM
| 1. | Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247-2250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1567] [Cited by in F6Publishing: 1567] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 2. | Iredale JP. Hepatic stellate cell behavior during resolution of liver injury. Semin Liver Dis. 2001;21:427-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 246] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 3. | Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21:437-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 373] [Cited by in F6Publishing: 385] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 4. | Reeves HL, Friedman SL. Activation of hepatic stellate cells--a key issue in liver fibrosis. Front Biosci. 2002;7:d808-d826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 353] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 5. | Wang BE, Wang TL, Jia JD, Ma H, Duan ZP, Li XM, Li J, Wang AM, Qian LX. Experiment and clinical study on inhibition and reversion of liver fibrosis with integrated Chinese and Western Medicine. CJIM. 1999;5:6-11. [Cited in This Article: ] |
| 6. | Yin SS, Wang BE, Wang TL, Jia JD, Qian LX. [The effect of Cpd 861 on chronic hepatitis B related fibrosis and early cirrhosis: a randomized, double blind, placebo controlled clinical trial]. Zhonghua Ganzangbing Zazhi. 2004;12:467-470. [PubMed] [Cited in This Article: ] |
| 7. | Wang TL, Wang BE, Zhang HH, Liu X, Duan ZP, Zhang J, Ma H, Li XM, Li NZ. Pathological study of the therapeutic effect on HBV –related liver fibrosis with herbal compound 861. Weichangbingxue He Ganbingxue Zazhi. 1998;7:148-153. [Cited in This Article: ] |
| 8. | Taimr P, Higuchi H, Kocova E, Rippe RA, Friedman S, Gores GJ. Activated stellate cells express the TRAIL receptor-2/death receptor-5 and undergo TRAIL-mediated apoptosis. Hepatology. 2003;37:87-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 165] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 9. | Cao Q, Mak KM, Ren C, Lieber CS. Leptin stimulates tissue inhibitor of metalloproteinase-1 in human hepatic stellate cells: respective roles of the JAK/STAT and JAK-mediated H2O2-dependant MAPK pathways. J Biol Chem. 2004;279:4292-4304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 139] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 10. | Sutherland MW, Learmonth BA. The tetrazolium dyes MTS and XTT provide new quantitative assays for superoxide and superoxide dismutase. Free Radic Res. 1997;27:283-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 175] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 11. | Dunigan DD, Waters SB, Owen TC. Aqueous soluble tetrazolium/formazan MTS as an indicator of NADH- and NADPH-dependent dehydrogenase activity. Biotechniques. 1995;19:640-649. [PubMed] [Cited in This Article: ] |
| 12. | Stagliano KE, Carchman E, Deb S. Real-time polymerase chain reaction quantitation of relative expression of genes modulated by p53 using SYBR Green I. Methods Mol Biol. 2003;234:73-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Zhang FK, Wang BE, Wang TL, Jia JD, Dong Z, Zhang J. Effect of different pharmaceutics of herbal compound 861 on experi-mental liver fibrosis. Zhonghuo Zhongxiyi Jiehe Zazhi. 2000;10:15-16. [Cited in This Article: ] |
| 14. | Jia JD, Wang BE, Dong Z, Cui L, Zhu JX, Che JT. The effect of herbal compound 861 on mRNA levels for type I, III, and IV col-lagens and TGF-β in immune complex rat liver fibrosis. Zhonghua Ganzangbing Zazhi. 1996;4:214-216. [Cited in This Article: ] |
| 15. | Ma H, Wang BE, Ma XM, Jia JD. Effect of herbal compound 861 on rat hepatic stellate cell collagen synthesis and degrada-tion in vitro. Zhonghua Ganzangbing Zazhi. 1999;7:30-32. [Cited in This Article: ] |
| 16. | Ding HG, Tang SZ, Wang BE, Jia JD, Zhao CH. [Effects of herbal compound 861 on hepatic stellate cell expressing endothelin-1 protein and mRNA]. Zhonghua Ganzangbing Zazhi. 2003;11:308. [PubMed] [Cited in This Article: ] |
| 17. | Yin C, Ma H, Wang A, Ma X, Jia J, Wang B. [Effect of compound 861 on tissue inhibitor of metalloprotenase 1 gene expression of HSC-T6 cells]. Zhonghua Ganzangbing Zazhi. 2002;10:197-199. [PubMed] [Cited in This Article: ] |
| 18. | Ding HG, Wang BE, Shang HW. [Effect of herbal compound 861 on expression and activity of nitric oxide synthase in hepatic stellate cells]. Zhongguo Zhongxiyi Jiehe Zazhi. 2002;22:362-364. [PubMed] [Cited in This Article: ] |
| 19. | Yin CH, Ma H, Wang AM, Ma XM, Jia JD, Wang BE. Effect of compound 861 on stromelysin gene expression of HSC-T6 cell. Linchuang Ganzangbing Zazhi. 2002;18:168-170. [Cited in This Article: ] |
| 20. | You H, Wang B, Wang T. [Proliferation and apoptosis of hepatic stellate cells and effects of compound 861 on liver fibrosis]. Zhonghua Ganzangbing Zazhi. 2000;8:78-80. [PubMed] [Cited in This Article: ] |
| 21. | Ma H, Wang BE, Ma XM, Jia JD. Effect of compound 861 on he-patic stellate cell proliferation and collagen synthesis. Zhongguo Linchuang Yaolixue Yu Zhiliaoxue Zazhi. 1998;3:172-175. [Cited in This Article: ] |
| 22. | Wall SJ, Edwards DR. Quantitative reverse transcription-polymerase chain reaction (RT-PCR): a comparison of primer-dropping, competitive, and real-time RT-PCRs. Anal Biochem. 2002;300:269-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Pérez C, Vandesompele J, Vandenbroucke I, Holtappels G, Speleman F, Gevaert P, Van Cauwenberge P, Bachert C. Quantitative real time polymerase chain reaction for measurement of human interleukin-5 receptor alpha spliced isoforms mRNA. BMC Biotechnol. 2003;3:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 874] [Cited by in F6Publishing: 801] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 25. | Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 221] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Ramos-Payán R, Aguilar-Medina M, Estrada-Parra S, González-Y-Merchand JA, Favila-Castillo L, Monroy-Ostria A, Estrada-Garcia IC. Quantification of cytokine gene expression using an economical real-time polymerase chain reaction method based on SYBR Green I. Scand J Immunol. 2003;57:439-445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |