Published online Nov 7, 2017. doi: 10.3748/wjg.v23.i41.7440
Peer-review started: June 2, 2017
First decision: July 27, 2017
Revised: August 23, 2017
Accepted: September 20, 2017
Article in press: September 19, 2017
Published online: November 7, 2017
To identify circulating micro (mi)RNAs as biological markers for prediction of severe acute pancreatitis (SAP) with acute lung injury (ALI).
Twenty-four serum samples were respectively collected and classified as SAP associated with ALI and SAP without ALI, and the miRNA expression profiles were determined by microarray analysis. These miRNAs were validated by quantitative reverse transcription-polymerase chain reaction, and their putative targets were predicted by the online software TargetScan, miRanda and PicTar database. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (commonly known as KEGG) were used to predict their possible functions and pathways involved.
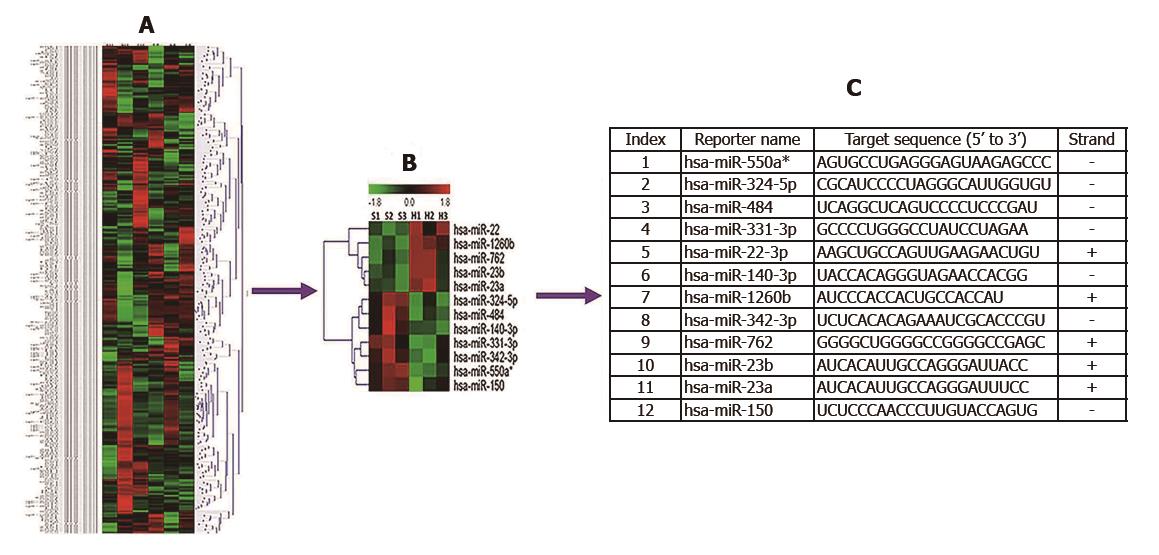
We investigated 287 miRNAs based on microarray data analysis. Twelve miRNAs were differentially expressed in the patients with SAP with ALI and those with SAP without ALI. Hsa-miR-1260b, 762, 22-3p, 23b and 23a were differently up-regulated and hsa-miR-550a*, 324-5p, 484, 331-3p, 140-3p, 342-3p and 150 were differently down-regulated in patients with SAP with ALI compared to those with SAP without ALI. In addition, 85 putative target genes of the significantly dysregulated miRNAs were found by TargetScan, miRanda and PicTar. Finally, GO and pathway network analysis showed that they were mainly enriched in signal transduction, metabolic processes, cytoplasm and cell membranes.
This is the first study to identify 12 circulating miRNAs in patients with SAP with ALI, which may be biomarkers for prediction of ALI after SAP.
Core tip: Early diagnosis of severe acute pancreatitis (SAP) associated with acute lung injury (ALI) is still difficult. Our study is the first to identify 12 differentially expressed circulating microRNAs in patients with SAP with ALI, which may be used as circulating biomarkers for prediction of acute lung injury induced by SAP.
- Citation: Lu XG, Kang X, Zhan LB, Kang LM, Fan ZW, Bai LZ. Circulating miRNAs as biomarkers for severe acute pancreatitis associated with acute lung injury. World J Gastroenterol 2017; 23(41): 7440-7449
- URL: https://www.wjgnet.com/1007-9327/full/v23/i41/7440.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i41.7440
Acute pancreatitis is a potentially fatal disease characterized by wide clinical variation, ranging from a mild self-limiting to severe disease complicated with sepsis and multiple organ failure, leading to high morbidity and mortality[1]. Severe acute pancreatitis (SAP) develops in 10%-25% of the patients, which is also a systemic disease and usually induces injury to extra-pancreatic organs, such as lung, liver and kidney, leading to multiple organ failure and even death[2,3]. Acute lung injury (ALI) is a common and serious complication of SAP. It can develop into acute respiratory distress syndrome (ARDS) without timely intervention[4,5]. At present, the early diagnosis of SAP associated with ALI is still difficult. Some biochemical parameters, such as high-mobility group box (HMGB)-1, pre-B cell colony-enhancing factor (PBEF) and surfactant protein (SP)-A, have been used for diagnosis of ALI[6-9]. However, there are few research studies in this area and there are no definitive diagnostic markers; additionally, sometimes the concentration of the markers can be affected by other factors. So, it is important to choose several biomarkers that might be used jointly to improve the diagnostic rate and establish the mechanism of SAP associated with ALI.
Micro (mi)RNAs are a family of naturally occurring, small noncoding RNA molecules that play an important regulatory role in gene expression. They are involved in numerous pathophysiological processes within cells and represent major regulators of gene expression by virtue of their preponderance to target transcription factors. miRNAs have been proposed as ideal biomarkers of disease, including diagnosis, prognosis and monitoring of treatment responses. Circulating miRNAs, presenting in a stable form protected from endogenous RNase activation, are highly stable in blood serum. Hence, in view of their potential use as novel, noninvasive biomarkers, the profiles of circulating miRNAs have been explored in diseases such as cancer, heart failure, myocardial infarction and spinal cord injury[10-17]. miRNA analysis of plasma from patients with SAP associated with ALI might yield new biomarkers for diagnosis and identify potential treatment targets for ALI[18,19].
The present study was designed to test the hypotheses that circulating miRNAs are closely related to the incidence of SAP associated with ALI, they are be involved in a variety of pathophysiological processes of ALI, and can be used as a circulating marker to predict the disease. This study aimed to identify expression of the different circulating miRNAs and their target genes in patients with SAP associated with ALI, and in those with SAP without ALI. The findings from this study will contribute to early diagnosis and treatment of SAP associated with ALI.
All patients with SAP were admitted within 72 h after onset of symptoms to the Medical or Surgical Intensive Care Units of the Affiliated Zhongshan Hospital of Dalian University and Nanjing General Hospital of Nanjing Military Command from January 2010 to December 2013. Twelve of 30 SAP patients were not complicated with ALI. All patients met the criteria of the Atlanta definition of SAP. Specifically, SAP diagnosis should meet at least one of the following criteria: (1) APACHE II score ≥ 8; (2) Ranson score ≥ 3; (3) organ failure (i.e. transient and persistent); and (4) local complications (i.e. necrosis, abscess or pseudocyst). To ensure inclusion of only eligible patients with a first attack of SAP, patients with recurrent SAP, chronic pancreatitis or pancreatic cancer were excluded. ALI was defined by the American-European Consensus Conference[20].
Patient demographic and clinical characteristics are summarized in Table 1. Six SAP patients were finally excluded for not meeting all inclusion criteria, and 24 patients met the criteria; six of 24 serum samples of patients were randomly selected for miRNA microarray analysis. This study was approved by the Ethics Committee of the Affiliated Zhongshan Hospital of Dalian University and Nanjing General Hospital of Nanjing Military Command. All of the patients gave written informed consent.
| Characteristic | SAP with ALI cases | SAP without ALI cases | P value |
| Age | 43.85 ± 1.70 | 44.25 ± 1.47 | 0.8602 |
| Men/Women | 8/4 | 7/5 | 0.6733 |
| Current smoker | 6 (50) | 4 (33.3) | 0.6802 |
| Etiology | |||
| Biliary | 10 (83.3) | 9 (75) | 0.1606 |
| Alcohol | 1 (8.3) | 2 (16.7) | 0.1261 |
| Other | 1 (8.3) | 1 (8.3) | 1.0000 |
| APACHE II | 18.92 ± 1.00 | 13.17 ± 0.81 | 0.0002 |
| Mechanical ventilation | 8 | 0 | 0.0013 |
| Hospital mortality | 2 | 0 | 0.4783 |
Four-milliliter serum specimens from each patient were collected immediately after grouping of patients into SAP without ALI and SAP with ALI. Serum samples were immediately frozen in liquid nitrogen and then stored at -80 °C for later determination. Total RNA was extracted from serum specimens using a mirVana PARIS kit (Ambion, Austin, TX, United States). The quantity and quality of total RNA were determined using an ultraviolet spectrophotometer at 260 and 280 nm.
miRNA microarrays were manufactured by Agilent Technologies (Santa Clara, CA, United States). Total RNA samples (4-8 µg) were 3’-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining (Atactic Technologies, Houston, TX, United States)[21-23]. Similar methods reported in a previous work were used in this section[24,25]. All raw data were transformed to log2 values, and each expression was normalized by having zero mean and unit sample variance. In order to screen significantly differentially expressed miRNAs, we compared expression in serum between the patients with SAP with ALI and those with SAP without ALI.
The relative miRNA expression levels were further normalized using the overall median value for patients with SAP with ALI, which gave each patient a median log ratio of 0 for normalized expression levels. The weighted differences in miRNA expression between patients with SAP with ALI and those with SAP without ALI were calculated using the random variance model t-test, in which fold-change > 1.5 was considered significant. The heatmap analysis and hierarchical cluster analysis of data were performed using Multi-Experiment Viewer (MeV) v4.7.1 from the TM4 software package available as open-source software at http://www.tm4.org[26]. Hierarchical clustering was performed using the Euclidean distance metric with complete linkage option.
qRT-PCR was used to verify miRNA expression identified by microarray analysis. Of 24 serum samples of patients, 18 were assessed by qRT-PCR. The primers for RT-PCR were used to amplify miRNAs, and were designed using Premier 5.0.RNase inhibitor (Toyobo, Osaka, Japan). The reverse primer, M-MLV reverse transcriptase and U6B were purchased from Guangzhou RiboBio Co. Ltd. (China), and U6B was used as internal control. Mixtures of 1 μg total RNA, 50 nmol/L reverse primer, 5 U M-MLV reverse transcriptase (Toyobo), 2 U RNase inhibitor (Toyobo) and 0.5 μmol/L dNTP were incubated for 30 min at 16 °C, 30 min at 42 °C, and 15 min at 70 °C. The reaction mixtures were used as templates for qRT-PCR. PCR product amplification was determined by the level of fluorescence emitted by SYBR Green Realtime PCR Master Mix-Plus (Takara, Dalian, China) and MX3000P qPCR system (Stratagene, Santa Clara, CA, United States). The PCR mixture, containing 1 μL reverse transcriptase product of total RNA, 10 μL SYBR-Green Real-time PCR Master Mix-plus, 2 μL Plus solution, 2 μL each specific forward and reverse primer, and 3 μL diethyl pyrocarbonate water, was prepared in a total final volume of 20 μL. The reaction was performed at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Each sample was run in triplicate. Melting curves were used to verify the specificity of each PCR. The procedure for qRT-PCR was described previously[27].
The putative target prediction of validated miRNAs by qRT-PCR was performed using three online software programs: PicTar: http://www.pictar.org/; miRanda: http://www.microrna.org/microrna/home.do/; and TargetScan: http://www.targetscan.org. We used PicTar, miRanda and TargetScan together to enhance the credibility of the study. PicTar identified a list of putative targets, searching for almost fully complementary sites between miRNAs and 3’-untranslated region (UTR) mRNAs. The free energy between the binding sites was then calculated and the results were ranked by means of a score obtained using a hidden Markov model (HMM). miRNAs with multiple binding sites were highly scored. The miRanda algorithm searched for target sites on the 3’-UTRs of mRNAs. It considered both the binding energy for the duplex stability and the conservation of the target site among different species. The TargetScan algorithm was based on the identification of fully complementary zones between the miRNA seed (nucleotides 2-8) and 3’-UTR mRNA. Starting from those sites, TargetScan searched for larger interactions, ranking the results in three groups according to the length of the matches. In particular, the presence of an adenine in the first position of the target site was highly scored because of its evolutionary conservation. The prediction followed the rules: (1) perfect match at the seed region (2-8 nucleotides from the 5’ end of miRNA); (2) minimum free energy of the miRNA/target duplex should be smaller than -20 kcal/mol; and (3) total score for miRNA-mRNA pairs should be > 140. GO was performed for analysis of the biological process (BP), cellular component (CC) and molecular function (MF) of miRNA target genes based on the GO database (http://www.geneontology.org/). Fisher’s two-side exact test and χ2 test were used to classify the GO categories, and the false discovery rate (FDR) was calculated to correct the P values. We chose only GOs that had a P value < 0.05 and FDR < 0.05.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to identify the enriched pathways of miRNA target genes based on the KEGG database (http://www.genome.jp/kegg/). This analysis provided a better understanding of gene expression information as a complete network. KEGG pathway annotation of the miRNA targets was found using the Database for Annotation, Visualization and Integrated Discovery (DAVID) gene annotation tool (http://david.abcc. ncifcrf.gov/). Fisher’s exact test and the threshold of significance were defined by the P value and FDR. The screening criterion was P < 0.05. To calculate the enrichment ratio and P value for KEGG analysis, we defined N as number of genes annotated by pathway chip and M as number of differentially expressed genes annotated by pathway in predicted miRNA targets. We recorded the intersecting genes belonging to GO and the pathway at the same time. According to the attributes of the intersecting target genes and miRNAs, the miRNA gene network, representing the critical miRNAs and their targets, was established in accordance with the miRNA degree. The key miRNA and gene in the network had the largest extent. The most important biological metabolic pathway and signal transduction pathway could be determined using pathway enrichment.
Statistical analyses were performed using GraphPad Prism 6.0 software (San Diego, CA, United States). Fisher’s exact test and t-test were used to assess differences in clinical features between the participants in the two groups. A value of P < 0.05 was considered statistically significant.
We investigated 287 miRNAs based on microarray data analysis. The miRNAs with fold-changes > 1.5 or < 1/2 and P < 0.05 were selected for further study. Twelve miRNAs met these criteria and were differentially expressed in patients with SAP with ALI, and in those with SAP without ALI. The 12 miRNAs are summarized in Figure 1.
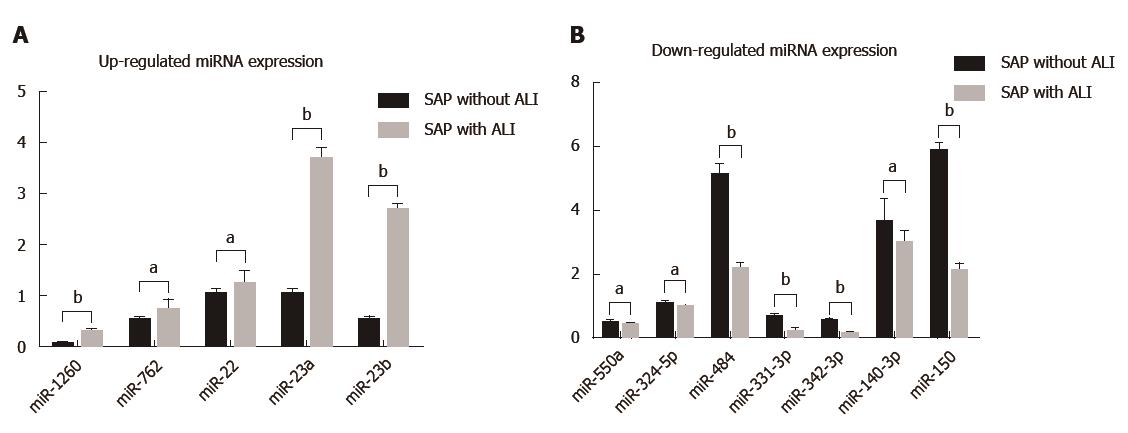
In order to validate the microarray results, qRT-PCR was performed for the identified 12 miRNAs (P < 0.05). We found that five miRNAs (hsa-miR-22-3p, hsa-miR-1260b, hsa-miR-762, hsa-miR-23b and hsa-miR-23a) were significantly up-regulated and seven (hsa-miR-550a*, hsa-miR-324-5p, hsa-miR-484, hsa-miR-331-3p, hsa-miR-140-3p, hsa-miR-342-3p and hsa-miR-150) were down-regulated in patients with SAP with ALI and in those with SAP without ALI (Figure 2).
Eighty-five putative target genes of the 12 validated miRNAs were found in patients with SAP with ALI and those with SAP without ALI by TargetScan, miRanda and PicTar database.
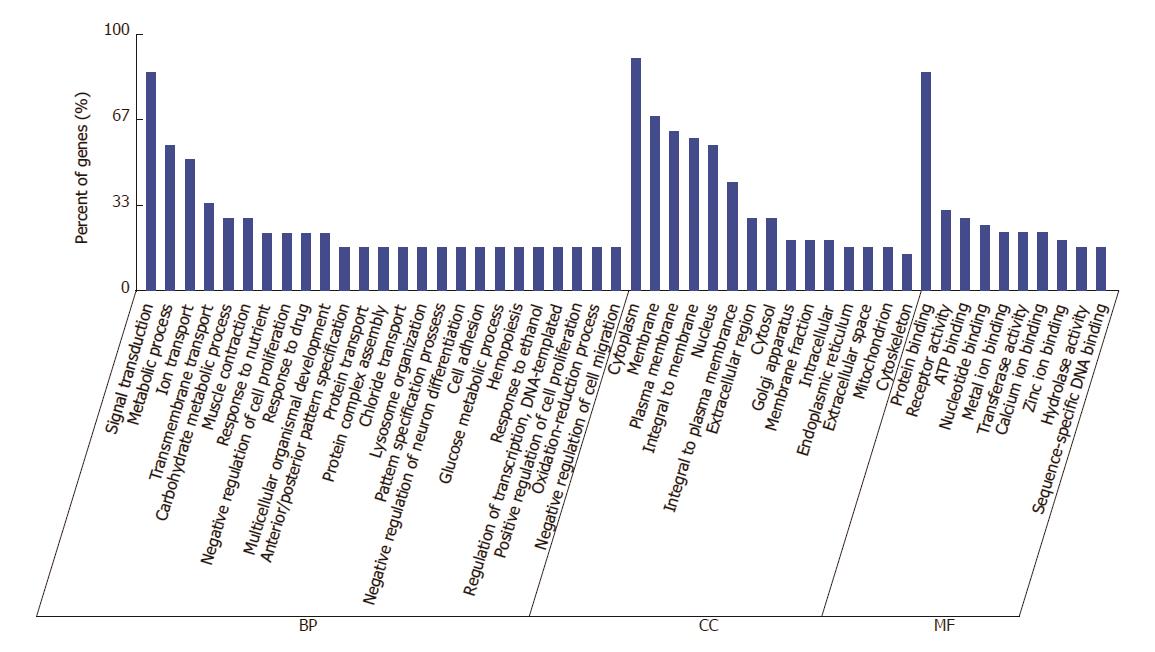
GO analysis showed annotation of the genes from three ontologies: BP, MF and CC. GO enrichment analysis of 12 validated miRNAs indicated that 4817 GOs were regulated by the down-regulated miRNAs, whereas 5088 GOs were regulated by the up-regulated miRNAs. In BP terms, they were enriched in signal transduction and metabolic processes. In CC terms, they focused on cytoplasm, cell membranes and plasma membranes, and were enriched in protein binding and in receptor activity terms in MF (Figure 3).
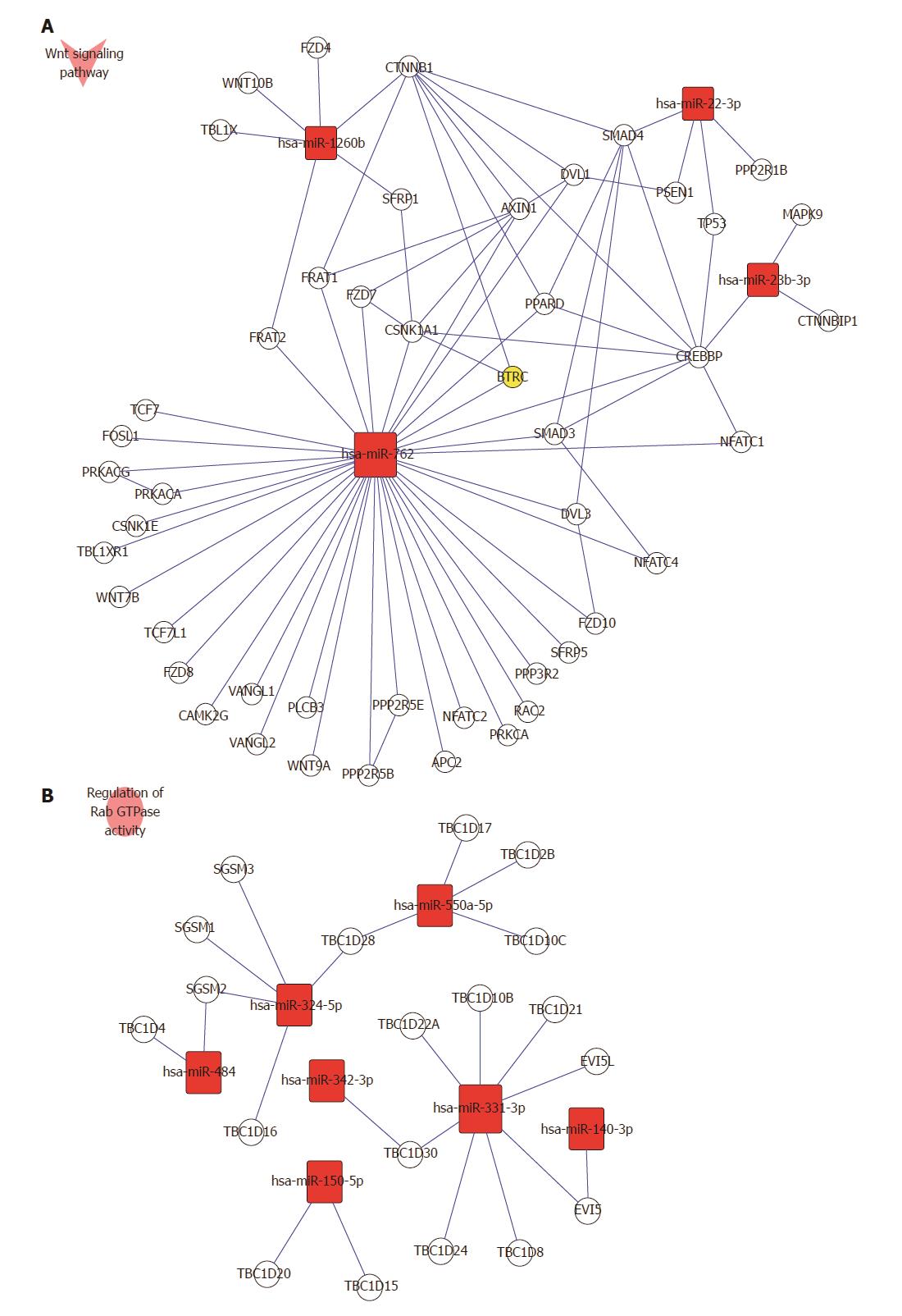
KEGG pathway analysis showed that the target genes of up-regulated miRNAs were involved in glutathione metabolism, Wnt signaling pathway, cytokine-cytokine receptor interaction, complement and coagulation cascades, methionine metabolism, apoptosis, cytosolic DNA-sensing pathway, Notch signaling pathway, chemokine signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway. Target genes of down-regulated miRNAs were related to the insulin signaling pathway, transforming growth factor (TGF)-β signaling pathway, T-cell receptor signaling pathway, amino sugar and nucleotide sugar metabolism, and starch and sucrose metabolism. As an example, the Wnt signaling pathway involved in up-regulated miRNAs and regulation of Rab GTPase activity by down-regulated miRNAs is shown in Figure 4A and B.
In this study, we identified that 12 miRNAs, including five up-regulated (has-miR-22-3p, 1260b, 762, 23b and 23a) and 7 down-regulated (has-miR-550a-5p, 324-5p, 484, 331-3p, 22-3p, 140-3p and 342-3p) were significantly expressed in patients with SAP with ALI and in those with SAP without ALI. Furthermore, we found that 4817 GOs regulated by the seven down-regulated miRNAs and 5088 GOs regulated by the five up-regulated miRNAs were involved in SAP with ALI. These miRNAs may be candidate novel biomarkers and were involved in many biological processes of SAP with ALI, such as signal transduction, metabolic processes, cytoplasm, cell membranes, and receptor activity terms. Moreover, the 85 putative target genes were regulated by 12 specific miRNAs, and the Wnt signaling pathway and Rab GTPase activity were important metabolic pathways that were up-regulated and down-regulated by miRNAs, respectively.
miRNAs are short non-coding RNAs involved in post-transcriptional regulation of gene expression. An increasing body of evidence indicates that miRNAs have investigated the potential involvement of miRNAs in ALI or ARDS. Liu et al[28] found that miRNA-200c-3p is crucial in ALI/ARDS and that the inhibition of miR-200c-3p ameliorated the ALI induced by H5N1 virus infection in vivo, indicating a potential therapeutic target. Vaporidi et al[29] investigated pulmonary miRNA profiling in a mouse model of high tidal volume ventilation (HVTV)-induced lung injury and results showed that of the 335 miRNAs examined, the expression of 50 miRNAs increased > 2-fold, expression of 15 miRNAs decreased by more than half and the miRNAs with the greatest increase in expression after 4 h of HVT were miR-7b, miR-189 and miR-223, whereas the miRNAs with the greatest decrease in expression were miR-503 and miR-211. However, circulating miRNAs as biomarkers of SAP with ALI are still unclear.
In our study, 12 circulating miRNAs were differentially expressed in patients with SAP with ALI and SAP without ALI. Among these, five miRNAs (hsa-miR-22-3p, 1260b, 762, 23b and 23a) were significantly up-regulated and seven (hsa-miR-550a*, 324-5p, 484, 331-3p, 22-3p, 140-3p and 342-3p) were down-regulated. qRT-PCR was performed to verify the gene expression levels of these regulated circulating miRNA. In the five differentially overexpressed miRNAs, the expression of hsa-miR-1260b, 23b and 23a showed a fold-change > 5 with P < 0.01, while hsa-miR-22-3p and 762 showed only a > 2 but < 5 fold-change < 5 with P < 0.05. Xu et al[30] found that overexpression of miR-1260b in non-small cell lung cancer (NSCLC) with lymph node metastasis can be regarded as a specific signature for early progression and prognosis of NSCLC.
Sarrion et al[31] reported that expression of miR23a was correlated with pulmonary function, and silencing of miR23a resulted in increased expression of PGC1α, as well as its well-known regulated genes CYC, SOD, NRF2 and HO1 in patients with idiopathic pulmonary arterial hypertension. Begum et al[32] found that over-expression of miR-23b in H1838 cells significantly increased cell proliferation, while inhibition of miR-23b in H1437 and H1944 cell lines significantly reduced cell doubling time. In addition, our miRNA network analysis predicted that these up-regulated miRNAs modulate glutathione metabolism, cytokine-cytokine receptor interaction, complement and coagulation cascades, methionine metabolism, apoptosis, cytosolic DNA-sensing pathway, Notch signaling pathway, chemokine signaling pathway, and MAPK signaling pathway.
Ell et al[33] found that the miRNA-23b/27b/24 cluster promoted breast cancer lung metastasis by targeting the metastasis-suppressive gene prosaposin. Thus, these results show that the significantly overexpressed hsa-miR-1260b, 23a and 23b may be important biomarkers of SAP with ALI. Nevertheless, there is a lack of relevant studies on their role of pathological processes in ALI induced by ASP. Moreover, there is still no report about the reports with hsa-miR-22-3p and hsa-miR-762 and lung diseases.
We also identified seven down-regulated miRNAs (hsa-miR-550a*, 324-5p, 484, 331-3p, 140-3p, 342-3p and 150) by microarray analysis in patients with SAP with ALI and SAP without ALI. Expression of hsa-miR-484, 331-3p, 342-3p and 150 showed a fold-change < 0.5 with P < 0.01. miRNA-550a acts as a pro-metastatic gene and directly targets cytoplasmic polyadenylation element binding protein 4 in hepatocellular carcinoma[34]. Moreover, the miR550a-5p/RNF43/Wnt signaling axis acts as a Brg-1 target for regulating colorectal cancer metastasis, and miR150 likely regulates miR124a expression and thus augments expression of inflammatory mediators in myeKlf2-/- macrophages[35,36]. Cai et al[37] found that miR-199a and miR-16 were the most significantly down-regulated miRNAs in lipopolysaccharide-induced mouse ALI.
In our study, miR-199a and miR-16 expression did not differ between patients with SAP with ALI and those with SAP without ALI. This discrepancy may have been caused by different inducing factors of ALI. In addition, the results of KEGG pathway enrichment analysis and miRNA-KEGG network showed that the target genes of down-regulated miRNAs were related to the Wnt signaling pathway, insulin signaling pathway, TGF-β signaling pathway, T-cell receptor signaling pathway, and amino sugar and nucleotide sugar metabolism. Previous studies have shown that signaling pathways are involved in the pathogenesis of ALI. Guo et al[38] reported that Wnt3a over-rode the effect of P2X7R on Wnt/β-catenin signaling to prevent death of alveolar epithelial type I cells and restrict the severity of ALI. Pittet et al[39] reported that TGFbeta was a critical mediator of acute lung injury. Mu et al[40] also reported that the TGF-β1/Smad signaling (p-Smad 2 and p-Smad 3) pathway was an important mechanism in the course of ALI.
In conclusion, we identified 12 significantly expressed circulating miRNAs in patients with SAP with ALI. These miRNAs may be specifically involved in the pathological mechanism of SAP with ALI. miRNAs and their target genes, as novel therapeutic targets, look promising. At present, the study regarding the mechanism of miRNAs in ALI remains at its nascent stage. miRNAs and their role of pathological processes in ALI induced by SAP remain to be elucidated. There were some limitations in this study. The main limitation was the small sample size. The results need to be verified in multicenter clinical trials. In addition, the molecular mechanism of circulating miRNAs regulating downstream signaling pathways should be further studied in patients with SAP with ALI.
Severe acute pancreatitis (SAP) is a potentially fatal disease characterized by wide clinical variation. Acute lung injury (ALI) is a common and serious complication of SAP. At present, the early diagnosis of SAP with ALI is still difficult. So, it is important to choose several biomarkers that may be used jointly to improve the diagnostic rate and establish the mechanism of SAP with ALI.
Some biochemical parameters have been used for diagnosis of ALI, but there are no definitive diagnostic markers and few research studies in this area. We identify circulating biomarkers for prediction of SAP with ALI.
The authors identify 12 significantly expressed circulating miRNA in SAP with ALI patients. The information on these miRNAs shed light on potential pathological mechanisms underlying SAP associated with ALI and open new doors for the development of circulating biomarkers for SAP associated with ALI.
This observational study contributes to finding relevant biomarkers for prediction of SAP with ALI.
ALI is defined as a syndrome of inflammation and increased permeability that is associated with a constellation of clinical, radiologic and physiologic abnormalities that cannot be explained by, but may coexist with, left atrial or pulmonary capillary hypertension.
SAP associated with ALI is a critical situation in clinic; therefore, it is important to find relevant biomarkers for detecting SAP associated with ALI in patients. In the present study, the authors demonstrated that 12 circulating microRNAs are associated with SAP patients with ALI. In conclusion, this observational study is novel, interesting and has potential clinical application.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Zaja I, Zhao J S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y
| 1. | Liu Y, Zhou D, Long FW, Chen KL, Yang HW, Lv ZY, Zhou B, Peng ZH, Sun XF, Li Y. Resolvin D1 protects against inflammation in experimental acute pancreatitis and associated lung injury. Am J Physiol Gastrointest Liver Physiol. 2016;310:G303-G309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Lytras D, Manes K, Triantopoulou C, Paraskeva C, Delis S, Avgerinos C, Dervenis C. Persistent early organ failure: defining the high-risk group of patients with severe acute pancreatitis? Pancreas. 2008;36:249-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Fedorkiv MB. [Prevention and correction of pulmonary complications for severe acute pancreatitis]. Klin Khir. 2015;22-24. [PubMed] [Cited in This Article: ] |
| 4. | Wei M, Gong YJ, Tu L, Li J, Liang YH, Zhang YH. Expression of phosphatidylinositol-3 kinase and effects of inhibitor Wortmannin on expression of tumor necrosis factor-α in severe acute pancreatitis associated with acute lung injury. World J Emerg Med. 2015;6:299-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | De Campos T, Deree J, Coimbra R. From acute pancreatitis to end-organ injury: mechanisms of acute lung injury. Surg Infect (Larchmt). 2007;8:107-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Li G, Zhou CL, Zhou QS, Zou HD. Galantamine protects against lipopolysaccharide-induced acute lung injury in rats. Braz J Med Biol Res. 2016;49:e5008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Lee KA, Gong MN. Pre-B-cell colony-enhancing factor and its clinical correlates with acute lung injury and sepsis. Chest. 2011;140:382-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | Hong SB, Huang Y, Moreno-Vinasco L, Sammani S, Moitra J, Barnard JW, Ma SF, Mirzapoiazova T, Evenoski C, Reeves RR. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med. 2008;178:605-617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Chen XY, Wang SM, Li N, Hu Y, Zhang Y, Xu JF, Li X, Ren J, Su B, Yuan WZ. Creation of lung-targeted dexamethasone immunoliposome and its therapeutic effect on bleomycin-induced lung injury in rats. PLoS One. 2013;8:e58275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3218] [Cited by in F6Publishing: 3388] [Article Influence: 211.8] [Reference Citation Analysis (0)] |
| 11. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5636] [Cited by in F6Publishing: 6065] [Article Influence: 379.1] [Reference Citation Analysis (0)] |
| 12. | Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1954] [Cited by in F6Publishing: 2008] [Article Influence: 154.5] [Reference Citation Analysis (0)] |
| 13. | Zhao DS, Chen Y, Jiang H, Lu JP, Zhang G, Geng J, Zhang Q, Shen JH, Zhou X, Zhu W. Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc Pathol. 2013;22:444-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Hsu A, Chen SJ, Chang YS, Chen HC, Chu PH. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed Res Int. 2014;2014:418628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 15. | Peng L, Chun-guang Q, Bei-fang L, Xue-zhi D, Zi-hao W, Yun-fu L, Yan-ping D, Yang-gui L, Wei-guo L, Tian-yong H. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol. 2014;9:89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Hachisuka S, Kamei N, Ujigo S, Miyaki S, Yasunaga Y, Ochi M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord. 2014;52:596-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Zhou X, Wen W, Shan X, Qian J, Li H, Jiang T, Wang W, Cheng W, Wang F, Qi L. MiR-28-3p as a potential plasma marker in diagnosis of pulmonary embolism. Thromb Res. 2016;138:91-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 18. | Wang JF, Zha YF, Li HW, Wang F, Bian Q, Lai XL, Yu G. Screening plasma miRNAs as biomarkers for renal ischemia-reperfusion injury in rats. Med Sci Monit. 2014;20:283-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Kinet V, Halkein J, Dirkx E, Windt LJ. Cardiovascular extracellular microRNAs: emerging diagnostic markers and mechanisms of cell-to-cell RNA communication. Front Genet. 2013;4:214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818-824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4145] [Cited by in F6Publishing: 4039] [Article Influence: 134.6] [Reference Citation Analysis (0)] |
| 21. | Beier M, Hoheisel JD. Analysis of DNA-microarrays produced by inverse in situ oligonucleotide synthesis. J Biotechnol. 2002;94:15-22. [PubMed] [Cited in This Article: ] |
| 22. | Gao X, Gulari E, Zhou X. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579-596. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 181] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 23. | Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, Srivannavit O, Gulari E, Gao X, Zhou X. microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol. 2007;382:287-312. [PubMed] [Cited in This Article: ] |
| 24. | Gui Z, Li S, Liu X, Xu B, Xu J. Oridonin alters the expression profiles of microRNAs in BxPC-3 human pancreatic cancer cells. BMC Complement Altern Med. 2015;15:117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Wang Y, Lan Q, Zhao X, Xu W, Li F, Wang Q, Chen R. Comparative Profiling of microRNA Expression in Soybean Seeds from Genetically Modified Plants and their Near-Isogenic Parental Lines. PLoS One. 2016;11:e0155896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374-378. [PubMed] [Cited in This Article: ] |
| 27. | Wu ZB, Li WQ, Lin SJ, Wang CD, Cai L, Lu JL, Chen YX, Su ZP, Shang HB, Yang WL. MicroRNA expression profile of bromocriptine-resistant prolactinomas. Mol Cell Endocrinol. 2014;395:10-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Liu Q, Du J, Yu X, Xu J, Huang F, Li X, Zhang C, Li X, Chang J, Shang D. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017;3:17021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 29. | Vaporidi K, Vergadi E, Kaniaris E, Hatziapostolou M, Lagoudaki E, Georgopoulos D, Zapol WM, Bloch KD, Iliopoulos D. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;303:L199-L207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Xu L, Li L, Li J, Li H, Shen Q, Ping J, Ma Z, Zhong J, Dai L. Overexpression of miR-1260b in Non-small Cell Lung Cancer is Associated with Lymph Node Metastasis. Aging Dis. 2015;6:478-485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Sarrion I, Milian L, Juan G, Ramon M, Furest I, Carda C, Cortijo Gimeno J, Mata Roig M. Role of circulating miRNAs as biomarkers in idiopathic pulmonary arterial hypertension: possible relevance of miR-23a. Oxid Med Cell Longev. 2015;2015:792846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 32. | Begum S, Hayashi M, Ogawa T, Jabboure FJ, Brait M, Izumchenko E, Tabak S, Ahrendt SA, Westra WH, Koch W. An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: Clinical significance of miR-23b-3p deregulation. Sci Rep. 2015;5:13236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 33. | Ell B, Qiu Q, Wei Y, Mercatali L, Ibrahim T, Amadori D, Kang Y. The microRNA-23b/27b/24 cluster promotes breast cancer lung metastasis by targeting metastasis-suppressive gene prosaposin. J Biol Chem. 2014;289:21888-21895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 34. | Tian Q, Liang L, Ding J, Zha R, Shi H, Wang Q, Huang S, Guo W, Ge C, Chen T. MicroRNA-550a acts as a pro-metastatic gene and directly targets cytoplasmic polyadenylation element-binding protein 4 in hepatocellular carcinoma. PLoS One. 2012;7:e48958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Wang G, Fu Y, Yang X, Luo X, Wang J, Gong J, Hu J. Brg-1 targeting of novel miR550a-5p/RNF43/Wnt signaling axis regulates colorectal cancer metastasis. Oncogene. 2016;35:651-661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 36. | Manoharan P, Basford JE, Pilcher-Roberts R, Neumann J, Hui DY, Lingrel JB. Reduced levels of microRNAs miR-124a and miR-150 are associated with increased proinflammatory mediator expression in Krüppel-like factor 2 (KLF2)-deficient macrophages. J Biol Chem. 2014;289:31638-31646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | Cai ZG, Zhang SM, Zhang Y, Zhou YY, Wu HB, Xu XP. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol. 2012;90:37-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 38. | Guo Y, Mishra A, Weng T, Chintagari NR, Wang Y, Zhao C, Huang C, Liu L. Wnt3a mitigates acute lung injury by reducing P2X7 receptor-mediated alveolar epithelial type I cell death. Cell Death Dis. 2014;5:e1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Pittet JF, Griffiths MJ, Geiser T, Kaminski N, Dalton SL, Huang X, Brown LA, Gotwals PJ, Koteliansky VE, Matthay MA. TGF-beta is a critical mediator of acute lung injury. J Clin Invest. 2001;107:1537-1544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 384] [Cited by in F6Publishing: 373] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 40. | Mu E, Ding R, An X, Li X, Chen S, Ma X. Heparin attenuates lipopolysaccharide-induced acute lung injury by inhibiting nitric oxide synthase and TGF-β/Smad signaling pathway. Thromb Res. 2012;129:479-485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |