Published online Oct 14, 2019. doi: 10.3748/wjg.v25.i38.5732
Peer-review started: May 5, 2019
First decision: June 10, 2019
Revised: July 26, 2019
Accepted: September 27, 2019
Article in press: September 27, 2019
Published online: October 14, 2019
In their seminal papers Hanahan and Weinberg described oncogenic processes a normal cell undergoes to be transformed into a cancer cell. The functions of ion channels in the gastrointestinal (GI) tract influence a variety of cellular processes, many of which overlap with these hallmarks of cancer. In this review we focus on the roles of the calcium (Ca2+), sodium (Na+), potassium (K+), chloride (Cl-) and zinc (Zn2+) transporters in GI cancer, with a special emphasis on the roles of the KCNQ1 K+ channel and CFTR Cl- channel in colorectal cancer (CRC). Ca2+ is a ubiquitous second messenger, serving as a signaling molecule for a variety of cellular processes such as control of the cell cycle, apoptosis, and migration. Various members of the TRP superfamily, including TRPM8, TRPM7, TRPM6 and TRPM2, have been implicated in GI cancers, especially through overexpression in pancreatic adenocarcinomas and down-regulation in colon cancer. Voltage-gated sodium channels (VGSCs) are classically associated with the initiation and conduction of action potentials in electrically excitable cells such as neurons and muscle cells. The VGSC NaV1.5 is abundantly expressed in human colorectal CRC cell lines as well as being highly expressed in primary CRC samples. Studies have demonstrated that conductance through NaV1.5 contributes significantly to CRC cell invasiveness and cancer progression. Zn2+ transporters of the ZIP/SLC39A and ZnT/SLC30A families are dysregulated in all major GI organ cancers, in particular, ZIP4 up-regulation in pancreatic cancer (PC). More than 70 K+ channel genes, clustered in four families, are found expressed in the GI tract, where they regulate a range of cellular processes, including gastrin secretion in the stomach and anion secretion and fluid balance in the intestinal tract. Several distinct types of K+ channels are found dysregulated in the GI tract. Notable are hERG1 upregulation in PC, gastric cancer (GC) and CRC, leading to enhanced cancer angiogenesis and invasion, and KCNQ1 down-regulation in CRC, where KCNQ1 expression is associated with enhanced disease-free survival in stage II, III, and IV disease. Cl- channels are critical for a range of cellular and tissue processes in the GI tract, especially fluid balance in the colon. Most notable is CFTR, whose deficiency leads to mucus blockage, microbial dysbiosis and inflammation in the intestinal tract. CFTR is a tumor suppressor in several GI cancers. Cystic fibrosis patients are at a significant risk for CRC and low levels of CFTR expression are associated with poor overall disease-free survival in sporadic CRC. Two other classes of chloride channels that are dysregulated in GI cancers are the chloride intracellular channels (CLIC1, 3 & 4) and the chloride channel accessory proteins (CLCA1,2,4). CLIC1 & 4 are upregulated in PC, GC, gallbladder cancer, and CRC, while the CLCA proteins have been reported to be down-regulated in CRC. In summary, it is clear, from the diverse influences of ion channels, that their aberrant expression and/or activity can contribute to malignant transformation and tumor progression. Further, because ion channels are often localized to the plasma membrane and subject to multiple layers of regulation, they represent promising clinical targets for therapeutic intervention including the repurposing of current drugs.
Core tip: Ion channels play an essential function in the physiology of the GI tract. There is increasing evidence that they are dysregulated at all stages of gastrointestinal (GI) cancer, from early initiation to metastasis. This information provides for the use of ion channel expression as useful clinical prognostic biomarkers in GI cancer. Perhaps more importantly new therapeutic modalities targeting ion channels in the GI tract, including the potential to target their dysregulation in GI cancers are becoming increasingly feasible. This strategy includes the repurposing of existing drugs that are used to treat other ion channel pathologies, or other diseases altogether. This review seeks to provide an overview of the role of ion channels in GI cancers with an emphasis on the potential for new therapies that target them.
- Citation: Anderson KJ, Cormier RT, Scott PM. Role of ion channels in gastrointestinal cancer. World J Gastroenterol 2019; 25(38): 5732-5772
- URL: https://www.wjgnet.com/1007-9327/full/v25/i38/5732.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i38.5732
In 2019, the American Cancer Society estimates that there will be more than 1.76 million new cases of cancer in the United States, accompanied by more than 607,000 cancer deaths[1]. Of these, the digestive system will have the highest number of new cases, and the second highest number of cancer deaths. The lifespan of cells in the gastrointestinal (GI) tract is very short. Propagated from stem cells, the epithelial cells of the stomach, small intestine, and colorectum are typically replaced in a matter of days and are some of the most replicative tissues in the body. This turnover is necessary due to the constant physical, chemical, and biological insults these tissues endure. This rapid proliferation increases the likelihood of cells in these tissues acquiring and accumulating oncogenic mutations.
The basic functions of the GI epithelium are: (1) to act as a physical barrier that selectively allows for the absorption of nutrients; while (2) excluding toxic or pathogenic substances; and (3) to excrete substances to aid in the digestion process. These functions require large quantities of water, ions, and nutrients to be transported across the epithelial layer. The significant driving force for this work is achieved through the use of ion gradients.
The unequal distribution of ions is required for the survival and function of any cell. This includes everything from concentration gradients across cellular and organellar membranes to gradients within the cytosol from one end to the other of a polarized cell. The distribution of ions is a consequence of the localization and activation of a variety of ion-specific channels, co-transporters, and pumps. Ion channels typically have a gating mechanism controlling when they are open or closed, and allow for the passive movement of select ions down their concentration gradient. In contrast to this passive dissipation of gradients, pumps make use of ATP hydrolysis to actively set up gradients. Co-transporters, or secondary pumps, exploit the energy gained by moving one ion down its gradient to power the movement of another ion or molecule against its gradient. Precise regulation of these elements in response to changing environmental conditions and the subsequent changes in ion concentration or flux are necessary for a multitude of cellular processes including proliferation, motility, absorption/secretion, apoptosis and many others. The goal of this review will be to summarize what we know about the role of ion channel dysfunction as it pertains to cancer development within the GI epithelium. We focus on the four main classes of ion channels: potassium (K+), chloride (Cl-), sodium (Na+), and calcium (Ca2+) and zinc (Zn2+) transporters and the major epithelial cancers that arise in the GI tract: colorectal cancer (CRC), gastric cancer (GC), esophageal cancer (EC), pancreatic cancer (PC) and hepatocellular carcinoma (HCC). For other important GI transporters and aquaporins and other types of cancers we refer readers to other specific reviews.
Nearly 20 years ago, Hanahan and Weinberg presented certain criteria that a normal cell must acquire to be transformed into a cancer cell[2]. These hallmarks of cancer have been expanded upon since that seminal paper, but their basic principles still remain[3]. The functions of ion channels influence a variety of cellular processes, many of which overlap heavily with these hallmarks of cancer. For this reason, cancer has been described as a channelopathy[4], with a recent review by Prevarskaya et al[5] asking whether cancer hallmarks are primarily oncochannelopathies. For example, Ca2+ channels play major roles in the control of cellular growth and proliferation, as well as the control of cell death[6]. K+ and Cl- channels are essential for the localized swelling and shrinking of different areas of a cell, necessary for cell migration[7]. Separate from their function as channels, studies of protein interactions with various ion channels have demonstrated their involvement in diverse processes such as cytoskeletal architecture and protein targeting[8]. It is clear, from the diverse influences of ion channels, that their aberrant expression and/or dysfunction can contribute to the transformation of normal cells into malignancy[4-5,9-15]. As discussed by Djamgoz et al[12] ion channels are expressed and dysregulated in all cancers throughout the multi-stage process, from initiation to metastasis. This is certainly true for the GI tract.
K+ channels play a major role in maintenance of plasma membrane (PM) potential. The action of the Na+/K+-ATPase transporter, H+/K+-ATPase transporter, and the NKCC cotransporter, coupled with the exit of K+ ions from the cell down their electrochemical gradient maintains a net intracellular negative charge at the PM. This hyperpolarized membrane potential is then used to drive the active transport of various molecules against their gradient. This is especially important in the GI epithelium, which must continuously transport mass quantities of water, electrolytes, and nutrients. With 77 genes coding for K+ channels, they are the largest, most diverse group of ion channels in the human genome. In addition, many of these genes have known splice variants, can be post-translationally modified, or form complexes with regulatory subunits. This immense variability in K+ channel function highlights the importance of being able to fine-tune K+ conductance and brings into question what other functions these channels might be serving. Generally, K+ channels are classified as either voltage-gated, Ca2+-activated, inward rectifier channels, or 2P-domain channels.
Whereas Nav channels are classically associated with the rapid depolarization of excitable cells, voltage-gated K+ (Kv) channels are responsible for re-polarization during an action potential. In non-excitable cells, such as those of the GI epithelium, the typical role of these channels is hyperpolarization of the PM. This negative membrane potential facilitates Ca2+ signaling and is required for regulation of intracellular pH and cell volume. Owing to these broad influences, K+ channels are implicated in a variety of cellular and tissue functions including cell proliferation and differentiation, pigmentation, hearing and the mammalian cochlea, contractility, circadian rhythms, migration, wound healing, cell cycle progression, apoptosis, autophagy, metabolism, angiogenesis, stem cell dynamics, and carcinogenesis, including cancer cell proliferation, invasion, migration and metastasis[9,11,16-20]. Notably, the mechanisms underlying loss of control of K+ channels are still not well understood[18].
Very prominent among the Kv channels contributing to cancer risk is KCNQ1, which demonstrates flexibility in gating permitting functional versatility[21]. In the GI tract KCNQ1 assembles with the β-subunits KCNE2 (gastric) or KCNE3 (intestinal) converting it from a voltage-dependent channel in the stomach into a voltage-independent, constitutively open channel in the intestine[22]. In gastric parietal cells, luminal KCNQ1/KCNE2 is essential for gastric acid secretion, working in conjunction with a H+/K+-ATPase. In the intestinal and colonic crypts, KCNQ1/KCNE3 is located basolaterally, and establishes the driving force for cAMP-mediated Cl- secretion through CFTR, necessary for mucus hydration[23,24]. In the colon, blocking the activity of KCNQ1/KCNE3 nearly entirely abolishes cAMP-mediated Cl- secretion, versus only about 50% in the small intestine, demonstrating a reliance on KCNQ1 in the colon[25].
Humans carrying germline mutations in KCNQ1 (Jervell and Lange-Nielsen and Romano-Ward syndromes) develop a range of pathologies, most notably cardiac arrhythmia (long and short QT), but also hearing loss, elevated gastrin levels, gastric hyperplasia and in some cases gastric neoplasia[26-30]. These phenotypes are well modeled in Kcnq1 knockout mice that develop inner ear defects, imbalance, chronic gastritis, gastric hyperplasia, and gastric metaplasia[31,32].
There is strong evidence for KCNQ1 functioning as a tumor suppressor in GI cancers. The first data came from Sleeping Beauty (SB) transposon mutagenesis screens for intestinal cancer in mice. Kcnq1 was the third-ranked common insertion site (CIS) gene (just behind Apc and Rspo2, well-known Wnt/β-catenin pathway signaling genes), with a predicted loss of function and 14 unique mutation sites, among 77 CIS genes identified in the first SB screen published in Science in 2009[33]. Kcnq1 was then identified as a CIS gene in three subsequent SB screens for intestinal cancer[34-36]. Kcnq1’s role was then confirmed in crosses of Kcnq1 knockout mice to the ApcMin mouse model of intestinal cancer where Kcnq1-deficiency significantly enhanced tumor phenotypes, including the development of invasive adenocarcinomas[37]. The role of KCNQ1 in human CRC was then investigated, finding that maintenance of KCNQ1 mRNA and protein levels was associated with significant disease-free and overall survival in stage II, III, and IV CRC[37,38]. Notably, for stage IV CRC patients following hepatic resections, maintenance of KCNQ1 protein expression conferred a 23-month survival advantage[37]. In other GI cancers Kcnq1 was a CIS gene in two SB screens for PC[39,40], one SB screen for HCC[41] and in one SB screen for GC, with a predicted loss of function[42]. Additional evidence in GC is provided by the phenotype of Kcnq1 knockout mice that develop gastric hyperplasia, metaplasia and occasional neoplasia[31,32] and in studies of human gastric cells where treatment of cells with atrial natriuretic peptide reduced cell proliferation by upregulating KCNQ1 expression[43]. In studies of HCC in human tissue and HCC cell lines, expression of KCNQ1 was down-regulated by promoter hypermethylation associated with epithelial to mesenchymal transition (EMT), and poor patient prognosis[44]. Additionally, in HCC it was reported that KCNQ1 regulated and sequestered β-catenin via physical interactions at the PM[44].
Although KCNQ1 deficiency is associated with poor outcome in CRC[37,38,45] and in HCC[44], the mechanisms underlying tumor suppression are not well understood. However, one clue is that KCNQ1 is localized to the base of the intestinal epithelial crypt which is the site of the stem cell compartment and the likely site of origin of CRC[46]. Functional significance of crypt localization was demonstrated by Than et al[37] who found that crypts isolated from KCNQ1-deficient colon epithelium displayed increased clonogenicity suggesting a possible selective advantage for tumor development. In addition, several studies demonstrate involvement of KCNQ1 with the Wnt/β-catenin pathway[38,44,45,47]. The Wnt/β-catenin pathway is vitally important in intestinal epithelial physiology and pathophysiology, with deregulation of the pathway contributing to over 80% of CRCs as well as a large percentage of HCCs. An early study in Xenopus oocytes demonstrated that β-catenin up regulated KCNQ1-mediated currents by promoting its insertion into PM without any effect on transcription[47]. More recent studies have looked at the interactions of KCNQ1 and β-catenin specifically in GI cancers. Analysis of 386 human stage II and III CRC tumors found correlation between KCNQ1 membrane-associated protein and nuclear β-catenin protein expression, which was surprising as KCNQ1-low expression was associated with poor outcome[38]. It was proposed that KCNQ1 may be down-regulated by promoter methylation in some cancers so that in the presence of β-catenin, as seen in most CRC, KCNQ1 would be upregulated, but if in addition the promoter becomes methylated KCNQ1 would be down-regulated. In contrast, a second study found that β-catenin directly negatively regulated KCNQ1 transcription in several CRC cell lines[45]. This group also found that KCNQ1 promoted cell membrane localization of β-catenin. This had the effect of limiting oncogenesis both by preventing nuclear localization of β-catenin and by maintaining adherens junctions that prevent EMT. A third study reported that in HCC cell lines, KCNQ1 expression was enhanced by treatment with a methyltransferase inhibiter suggesting that expression may be down-regulated by promoter methylation[44]. However, KCNQ1 appeared to sequester β-catenin at the cell membrane and to limit its transcriptional activity. The situation is further complicated by the lncRNA KCNQ1OT1, which through the recruitment of chromatin and DNA modifying proteins, silences multiple genes in the KCNQ1 region[48]. KCNQ1OT1 has been associated with poor patient survival in several GI cancers, including CRC[49], EC[50], and HCC[51]. KCNQ1OT1 itself is regulated by β-catenin. The Kugoh group has demonstrated that nuclear β-catenin activates the transcription of KCNQ1OT1 through a TCF-1 binding site within its promoter region[52]. These studies suggest that multiple factors affect the interactions between KCNQ1 and β-catenin, and that the balance of these factors differs among cancers. But apparently loss of KCNQ1 activity, through whatever means, can provide a selective advantage to the tumor, as each of these studies demonstrates that KCNQ1 is a tumor suppressor. Channel openers such as retigabine have been developed as treatments for diseases caused by KCNQ1-deficiency[53]. Thus, understanding the contribution of KCNQ1 to cancer progression could lead to new cancer therapeutic opportunities.
Given its role as the β-subunit in the KCNQ1/KCNE2 channel in gastric tissue it is not surprising that deficiency for KCNE2 contributes to human GC cell growth and progression. This was supported by studies in Kcne2 knockout mice that develop gastritis cystica profunda and neoplasia, pyloric polyadenomas and invasive adenocarcinomas, linked to upregulation of cyclin D1[54-58]. Kcne3 (intestinal β-subunit) knockout mice also partially phenocopied loss of KCNQ1, demonstrating a disruption in intestinal Cl- transport[46].
While KCNQ1 seems to have tumor-suppressive effects, another Kv channel, the human ether-a-go-go related gene 1 (hERG1) channel, has been implicated as an oncogene in various GI cancers including CRC[59-62], PC[62-66], EC[67-70], and GC[68,71-74]. In CRC, while hERG1 is not expressed in normal colonic mucosa, a distinct upregulation in expression is reported in adenocarcinomas, with the highest levels occurring in metastatic cancers, where hERG1 expression was associated with poor patient prognosis, and with little to no expression in adenomas. hERG1 expression levels and activity were positively correlated to cell migration using channel inhibitors and clones expressing various levels of the protein[59]. This role in cancer cell migration and invasion was later expanded upon with the discovery of hERG1’s interactions with β1-integrins in PM complexes. hERG1 was shown to modulate β1-integrin mediated VEGF-A secretion through the recruitment of PI3K and AKT[72]. In PC hERG1 over-expression was reported in 59% of tumors[66] where it promoted cancer cell migration via modulation of F-actin organization[64]. hERG1 expression was also associated with lymph node involvement, tumor grade, TNM stage and poor patient prognosis[66]. One report linked hERG1 to dysregulation of the EGFR signaling pathway. In EC, hERG1 expression was associated with progression from Barrett’s esophagus (BE) to EC, again associated with poor patient prognosis[70]. Finally, in GC, hERG1 over-expression was reported in 69% of cancers, where it promoted angiogenesis by mediating VEGF-A secretion via AKT-dependent regulation of HIF-1, and similarly, its expression was associated with poor patient prognosis[71-74].
Another ether-a-go-go family member, EAG1, is also reported to function as an oncogene in several GI cancers, including CRC[18,75,76], EC[75], and HCC[16,62,75,77]. In all cases EAG1 is reported to be over-expressed, and associated with invasion of cancer cells and poor patient prognosis.
At least nine other K+ channels have been reported to be dysregulated in GI cancers. Those acting as oncogenes include KCNA5 in GC[73,78-80], and CRC[79]; KCNC1 in CRC[76]; KCND1 in GC[81]; KCNJ3 in PC[65,82]; KCNN4 in CRC[5,14,83], PC[65,84], and HCC; KCNS3 in CRC[85]; and KCNK9[5,86,87] in CRC. Those acting as potential tumor suppressors include KCNA3 in CRC[4,76,88] and PC[65,82,89], and KCNH5 in EC[90]. Of note, the pathophysiological actions of many of these channels seem to involve mechanisms that are independent of their channel action. An example is KCNS3/Kv9.3, an electrically silent subunit in excitable cells, whose expression has been found increased in CRC and lung cancer cells, with a knockdown showing interference at the G0/G1 and G1/S cell cycle transitions[85]. Overall, the great majority of K+ channels discussed here function as oncogenes (with KCNQ1 a prominent exception), consistent with a model of K+ channel dysregulation/overexpression being necessary for cancer cell cycle and tumor progression. See Table 1 for a full listing of potassium ion channels and their role in GI cancers.
| Gene name (s) | Cancer | Role | Functional activity |
| KCNA3/Kv1.3 | Colorectal | Unclear | One report that Kv1.3 is frequently hypermethylated and expression down-regulated in CRC; a different report that Kv1.3 is upregulated in human and mouse colon carcinomas[4,76,88] |
| Pancreatic | Tumor suppressor | Expression down-regulated by promoter hypermethylation; promotes metastasis[65,82,89] | |
| KCNA5/Kv1.5 | Gastric | Oncogene | Expression up-regulated; silencing in GC cells inhibits proliferation; alters drug resistance[73,78-80] |
| Colorectal | Oncogene | Expression up-regulated[79] | |
| KCNC1/Kv3.1 | Colorectal | Oncogene | Expression up-regulated[76] |
| KCND1/Kv4.1 | Gastric | Oncogene | Expression up-regulated[81] |
| KCNE2/MiRP1 | Gastric | Tumor suppressor | Expression down-regulated; deficiency promotes tumor progression; knockout mice develop gastritis cystic profundal and neoplasia, pyloric polyadenomas; invasive adenocarcinomas; upregulation of cyclin D1; down-regulated in gastric cancer tissues and cell lines; overexpression in cell lines suppressed growth in soft agar and mouse tumor xenografts[54-58] |
| KCNH1/EAG1/Kv10.1 | Colorectal | Oncogene | Up-regulated; one report showed 75% of CRC tumors positive for Eag1; another report found overexpression in 3.4% of adenocarcinomas[18,75,76] |
| Esophageal | Oncogene | Expression up-regulated; associated with depth of invasion; independent negative prognostic factor[75] | |
| Hepatocellular | Oncogene | Expression up-regulated[16,62,75,77] | |
| KCNH2/hERG1/Kv11.1 | Colorectal | Oncogene | Expression up-regulated; triggers angiogenesis and tumor progression via inducement of PI3K/AKT signaling and HIF1-induced activation of VEGF-A; associated with invasiveness, poor prognosis for stage I and II; up-regulation in ApcMin and AOM mouse models enhanced cancer phenotypes[59-62] |
| Pancreatic | Oncogene | Expression up-regulated in 59% of pancreatic cancers; promotes migration of cancer cells by modulation of f-actin organization; associated with lymph node involvement, tumor grade, TNM stage, poor patient prognosis; linked to EGFR pathway; down-regulated by miR-96; overexpressed in pancreatic cancer cell lines[62-66] | |
| Esophageal | Oncogene | Expression upregulated; promotes progression from Barrett’s esophagus to esophageal cancer; associated with poor patient prognosis[67-70] | |
| Gastric | Oncogene | Expression up-regulated; stimulates angiogenesis by promoting VEGF-A secretion via AKT-dependent regulation of HIF1; positive in 69% of gastric cancers; associated with poor patient prognosis[62,71-74] | |
| KCNH5/EAG2/Kv10.2 | Esophageal | Tumor suppressor | Expression down-regulated by promoter hypermethylation[90] |
| KCNJ3/Kir3.1 | Pancreatic | Oncogene | Expression up-regulated[62,82] |
| KCNN4/Kca3.1 | Colorectal | Oncogene | Expression upregulated; promotes EMT[5,14,83] |
| Pancreatic | Oncogene | Expression up-regulated[65,84] | |
| Hepatocellular | Oncogene | Expression up-regulated[62] | |
| KCNS3/Kv9.3 | Colorectal | Oncogene | Silencing causes inhibition of proliferation of HCT15 CRC cells[85] |
| KCNK9/K2p9.1/Task3 | Colorectal | Oncogene | Expression up-regulated[5,86,87] |
| KCNN3/Kca2.3/SK3 | Colorectal | Oncogene | Expression and activity up-regulated; regulated by SigmaR1; physically coupled to Orai1[5,91] |
| KCNQ1/KvLqt1 | Colorectal | Tumor suppressor | Identified as a top 10 common insertion site (CIS) gene in a sleeping beauty transposon mutagenesis screen in mice; predicted loss of function in the screen; knockout mouse developed enhanced GI cancer phenotype in ApcMin model; expression down-regulated in human colorectal cancer, associated with poor prognosis in stage II, III, and IV disease; found to be down-regulated by β- catenin, which promotes EMT; in turn, KCNQ1 physically interacts with β-catenin, sequestering β-catenin at the plasma membrane[33-38,45] |
| Pancreatic | Not determined | Identified as a common insertion site (CIS) gene in two sleeping beauty transposon mutagenesis screens in mice[39,40] | |
| Gastric | Tumor suppressor | Identified as a CIS gene in a Sleeping Beauty transposon mutagenesis screen; predicted loss of function; knockout mouse susceptible to chronic gastritis, hyperplasia and metaplasia; atrial natriuretic peptide reduced proliferation of gastric cancer cells by upregulating KCNQ1[31,32,42,43] | |
| Hepatocellular | Tumor suppressor | Expression down-regulated by promoter hypermethylation; associated with poor patient prognosis; KCNQ1 regulated EMT; KCNQ1 regulates β-catenin physical interactions at the plasma membrane[41,44] | |
| KCNQ1OT1 | Colorectal | Oncogene | Expression up-regulated; promotes Wnt/β-catenin signaling and migration, poor patient prognosis[49,52] |
| Esophageal | Oncogene | Expression up-regulated; promotion of metastasis; poor patient prognosis[50] | |
| Hepatocellular | Oncogene | Expression up-regulated; competes with endogenous miR-504; promotes cell proliferation, associated with TNM stage and poor survival[51] |
The functions of Cl- channels in the GI tract include regulation of transepithelial transport (in particular major anions), volume control (osmoregulation), membrane potentials, lipid homeostasis, cell polarity, glucose and other metabolism, oxidative stress, inflammation, mucus, alterations in the microbiome, pH, cell motility, autophagy, mitochondrial dysfunction, apoptosis, cell polarity, cell-cell contact, stem cell function, and cellular immune responses[5,91-105]. All of these functions can be dysregulated in and contribute to malignant transformation, particularly in the GI tract due to its constant exposure to environmental influences.
CFTR encodes a Cl-, HCO3- anion channel found primarily on the apical surfaces of luminal epithelial cells. Mutations in CFTR are the cause of the hereditary life shortening disease cystic fibrosis (CF). Recently CFTR has been shown to be a tumor suppressor in CRC and CFTR deficiency has been implicated in several other cancers. Because the causal connection between CFTR and cancer has been most strongly established for CRC, this section of the review will focus primarily on CRC.
The CFTR gene found on chromosome 7 encodes an mRNA of 6128 nucleotides[106]. The CFTR protein consists of two symmetrical halves. Each half contains 6 membrane spanning domains joined by an intracellular regulatory region. The membrane spanning domains assemble to form an aqueous pore that allows flow of Cl- and HCO3- ions down their electrochemical gradients. In the intestine the flow of ions is from the cytoplasm to lumen. Ion specificity is provided by the amino acids lining the pore. Opening and closing is regulated by binding of ATP to two nucleotide binding domains in the regulatory region. ATP binding is mediated by cAMP activation of PKA which phosphorylates a site to open up the ATP binding domains. In vivo cAMP activity is commonly regulated by cholinergic stimuli. Outflow of ions creates osmotic pressure for the flow of water in the same direction so that CFTR indirectly determines the flow of water as well[107,108]. CFTR also regulates other ion channels (Na+, K+, Ca2+, and other Cl- channels). For example, CFTR indirectly regulates the cellular ionic environment by inhibiting activity of the Na+ importing channel SCNN1, which has the effect of further encouraging outflow of water and also by supporting additional HCO3- transporters[109]. CFTR also interacts with other membrane proteins to maintain epithelial tight junctions and barriers to fluid flow, adjusts the levels of acidity in secretions, and participates in the transport of sphingosine-1 phosphate, a regulator of cell adhesion and a signaling molecule for inflammation[99]. CFTR also contains a cytoplasmic C-terminal PDZ-binding motif. This domain interacts with PDZ-containing proteins involved in regulating the actin cytoskeleton and intracellular signaling[110,111]. CFTR is expressed on the apical surface of the intestinal epithelium throughout the length of the intestine. In the small intestine CFTR expression decreases from duodenum to ileum with strongest expression in the crypts[112]. In addition, functional CFTR is found on the brush border of villus cells and rare CFTR-high expressing cells throughout the small intestine[113]. In the colon CFTR is expressed in a proximal to distal gradient with highest concentration in the proximal region and cecum. CFTR is most highly expressed at the base of the crypt where intestinal epithelial stem cells reside[112,114]. Overall, CFTR is a major determinant of ion and water homeostasis and because it is highly expressed at the base of the crypt it is in a position to influence the intestinal stem cell compartment.
Inactivating germline mutations in CFTR cause the hereditary life shortening disease CF. CF is the most common autosomal recessive hereditary disease among Caucasians[115]. The most severe clinical manifestations are pulmonary inflammation and obstruction leading ultimately to pulmonary failure[116]. However, CFTR is expressed in many extra-pulmonary tissues including the linings of the pancreatic and biliary ducts where its loss ultimately leads to CF-related diabetes and CF-related liver disease[117,118]. CFTR dysfunction in the exocrine pancreas results in ion transport defects, β-islet cell-related disorders such as dysregulation of insulin release, obstruction of the pancreatic duct, chronic inflammation and cancer initiation[99,119]. CF patients also demonstrate defects in the male reproductive system, chronic sinusitis, and kidney stones. Older CF patients have dysregulation of glucose metabolism and sleep disorders caused by disruption of circadian rhythms[99]. CFTR is also highly expressed in the intestinal epithelium. Loss of CFTR in the intestine causes intestinal obstruction in the ileum and proximal colon known as meconium ileus in infants and distal intestinal obstruction in older patients. CF results in impaired absorption of nutrients due to pancreatic enzyme deficiency and possibly defects in lipid absorption[120]. CF patients are also prone to celiac disease, an intestinal disorder caused by gluten-mediated triggering of TH1 immune and antibody responses[102]. These and other clinical manifestations of CF demonstrate the profound importance of CFTR activity in many tissues.
The lifespan of individuals with CF has increased dramatically so that the average life expectancy of a person born with CF today is approximately 44 years[121]. As individuals with CF live longer it has become apparent that they are at increased risk for developing some but not all cancers. Initial evidence linking CF to cancer risk came from a 20-year epidemiological study that compared incidence of cancers in individuals in the United States Cystic Fibrosis registry to the predicted age adjusted risk in the general population to determine standardized incidence ratios. This study reported that the overall risk of cancer was not increased. However, the risk of all types of GI cancers was increased and in particular the risk of CRC, the most common GI cancer, was increased by 6-fold[122]. A recent meta-analysis of population-based studies is consistent with this result[123]. In support of the clinical significance of this finding, endoscopic screening studies of adult CF patients found that polyps in individuals with CF appeared earlier, and were larger and more aggressive than those in the non-CF population[124,125]. As a result of these studies the guidelines for endoscopic screening of CF patients have been modified and CF has been declared a hereditary colon cancer syndrome by the Cystic Fibrosis Foundation[126].
Mouse genetic studies demonstrated the functional significance of Cftr deficiency. SB transposon-mediated genetic screens initially identified Cftr as a candidate cancer-causing gene. Cftr was identified in the top 10% to 50% of candidate genes in three SB screens to identify CRC driver genes in Apc wildtype[33], Apc-deficient[34], and TGF beta-deficient[36] backgrounds[127]. Follow up studies evaluated mice carrying a targeted intestinal specific deletion of Cftr (Cftr-/-) for intestinal tumorigenesis. In the tumor sensitized ApcMin background Cftr-/- mice developed significantly more adenomas than ApcMinCftr wildtype mice. Further, ApcMinCftr-/- but not ApcMinCftr wildtype mice developed invasive lesions. Most striking, Cftr-deficiency alone was sufficient to cause adenomas in > 60% of mice after a one year interval[128].
CFTR deficiency is linked to CRC in the general population as well. In a study of 90 Stage II CRC patients stratified by tumor CFTR expression, disease-free survival at 3 years in the 25% of patients with lowest CFTR expression was 30% lower than those with higher expression[128]. In a second cohort, CFTR mRNA and protein expression was lower in tumor vs normal tissue and CFTR mRNA expression was lower in metastatic vs non-metastatic tumors. In this study, CFTR-depleted CRC cell lines showed enhanced oncogenic characteristics including increased colony formation, migration and invasion[129]. RAS, PKC and IFNα have been reported to be involved in the down-regulation of CFTR in the colon[119]. It is also possible that low CFTR expression in sporadic tumors is caused by epigenetic silencing of the CFTR promoter as has been reported in other cancers such as lung and bladder[130-133].
The single cell layer of the intestinal epithelium is replaced approximately every 5 days. Stem cells at the base of the crypt drive this renewal[114]. These stem cells and progenitor cells that acquire stem-like characteristics are thought to be the CRC initiating cells[134]. In the colon, CFTR expression is highest at the base of the crypt[112] and expression has been reported in the stem cell itself[93]. CFTR has also been reported to be involved in intestinal lineage differentiation with its knockdown causing proliferation, migration and expression of EMT genes[135]. Thus, CFTR is in a position to influence the renewal process of the intestinal epithelium and CFTR deficiency may directly influence cancer initiating cells. Intestinal crypts can be cultured in vitro as 3D organoid cultures. Organoid cultures recapitulate the intestinal luminal epithelial structure as well as the renewal and differentiation process[136]. CFTR is expressed in these cultures and maintains its ion channel function[137,138]. Thus, organoid cultures have been developed as surrogates to test the effectiveness of CF therapeutics as treatments for specific mutations[139-141]. In addition, organoid cultures derived from oncogenic or pre-oncogenic tissues maintain the oncogenic characteristics of these tissues[142,143]. Accordingly, organoid cultures have been used to determine the oncogenic characteristics of CFTR deficiency. Than et al[128] determined that Cftr-deficient colon organoids demonstrate increased clonogenicity. Analysis of small intestinal organoids by Strubberg et al[93] demonstrated increased proliferation of Cftr-deficient organoids and localization of Cftr to the LGR5 stem cell. These findings support a role for Cftr deficiency in the environment of the cancer initiating cell or the stem cell itself.
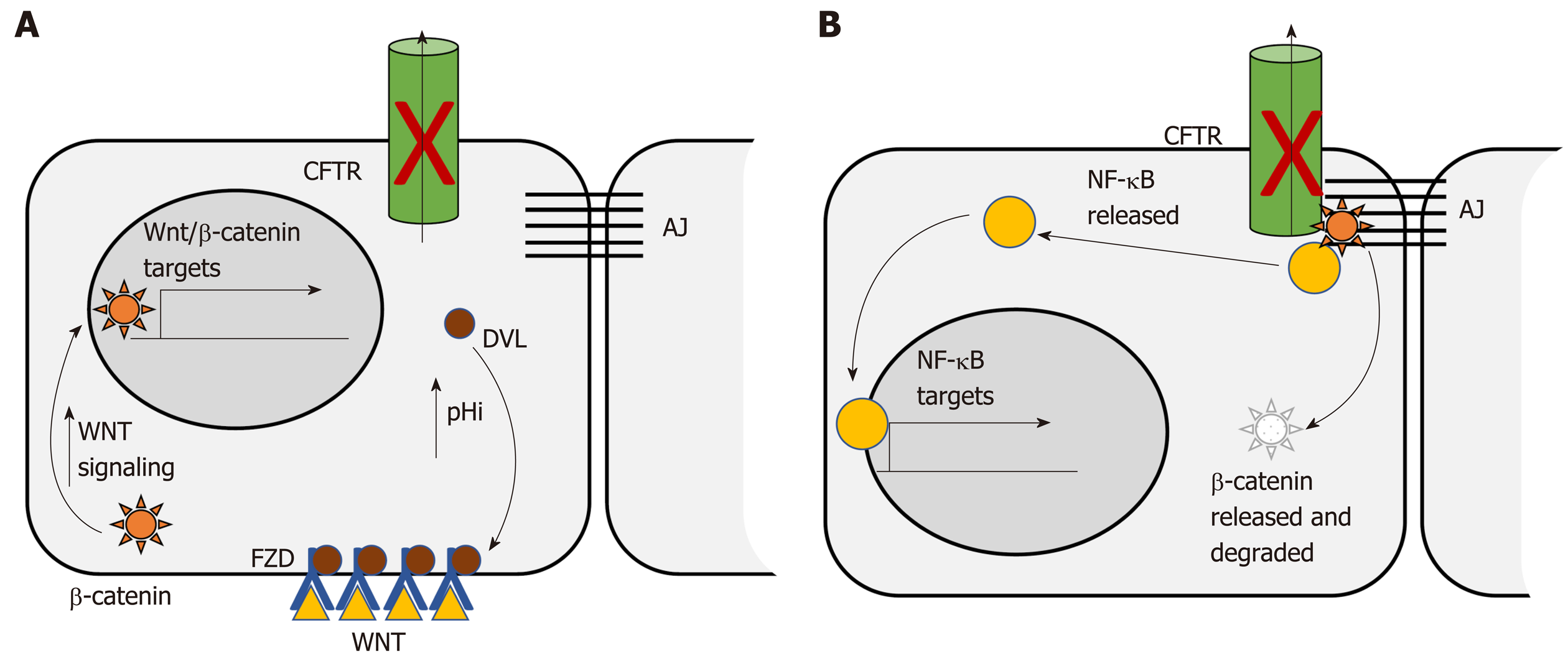
The Wnt/β-catenin signaling pathway is dysregulated in more than 85% of human CRC. Dysregulation contributes to initial adenoma formation and to maintenance of invasive tumors[144]. In the intestine CFTR deficiency is primarily associated with enhanced Wnt/β-catenin signaling. Than et al[128] identified nuclear localization of β-catenin, indicative of activation, as well as increased expression of Wnt/β-catenin target genes, in intestinal tumors deficient only for Cftr. Strubberg et al[93] identified increased active β-catenin in Cftr KO crypts and organoids which correlated with increased proliferation. Mechanistically, Cftr KO or transient inhibition of channel activity by CFTR(inh)-172 resulted in increased intracellular pH in LGR5+ stem cells. Increased pH in turn promoted association of Dvl2, a member of the Wnt/β-catenin signaling pathway, with PM phospholipids, which positioned it to enhance Wnt/β-catenin signaling[93] (Figure 1A). In contrast, Chan and colleagues reported that intestinal Cftr deficiency in mice carrying the F508del mutation and knockdown of CFTR in the Caco-2 CRC cell line results in diminished β-catenin signaling. Mechanistically, loss of CFTR destabilizes β-catenin membrane localization allowing it to be degraded leading to oncogenic phenotypes via activation of NF-κB[145] (Figure 1B). The differences between these two studies may reflect tissue specificity. This study analyzed Wnt/β-catenin signaling in total intestinal tissue rather than the crypt and so may reflect a different role for CFTR outside the stem cell compartment. Further, the effect of CFTR deficiency on Wnt/β-catenin activity varied by tissue in several other studies by this group[100]. In addition, Wnt/β-catenin signaling may also play different roles at different stages of CRC. Although it is generally accepted that dysregulated Wnt/β-catenin signaling is a driving force for initiation of CRC and progression[144] there is evidence that low levels of expression of Wnt/β-catenin targets are associated with poor prognosis in CRC[146]. Further, inhibition of Wnt/β-catenin has been shown to be necessary for survival of latent metastatic cells[147].
Extensive evidence that CFTR deficiency plays a role in many types of cancers suggests that tumor suppression by CFTR goes beyond regulation of Wnt/β-catenin signaling. The mechanisms of tumor suppression by CFTR are not well understood. However, the roles of CFTR in normal issue and the consequences of CF suggest plausible mechanisms.
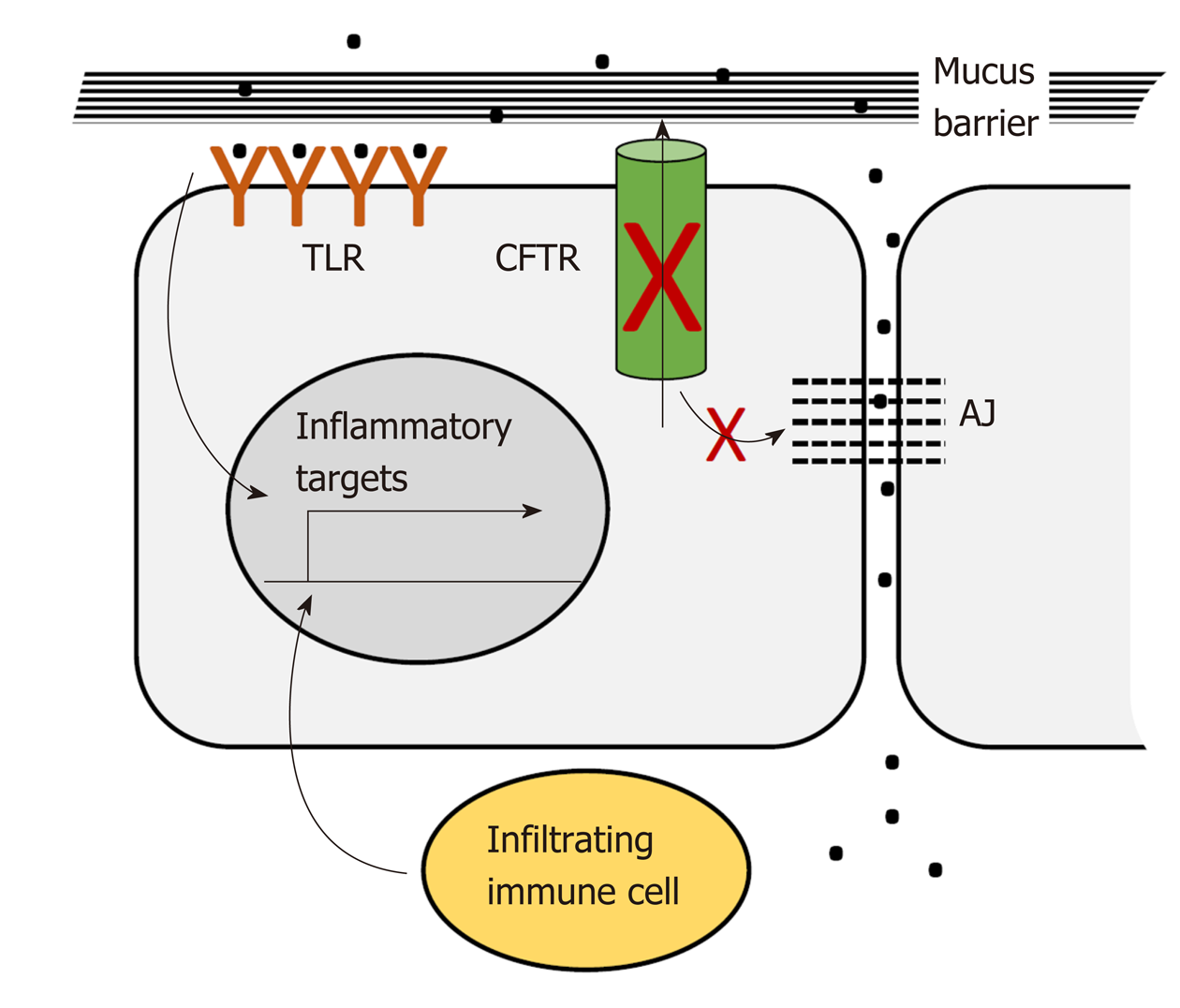
CFTR plays a pervasive role in intestinal homeostasis through its influence on several inter-related processes: Epithelial barrier maintenance, microbiota composition and immune homeostasis. Breakdown of these processes is responsible for some of the clinical manifestations of CF and is also potentially oncogenic (Figure 2).
The intestinal lumen is home to trillions of bacteria. These bacteria provide essential nutrients and signals needed for intestinal epithelial and immune cell homeostasis. However, direct contact with the intestinal epithelium must be prevented. The colon epithelium is protected from direct contact by protective mucus layers and by tight junctions between the lateral surfaces of the single layer of epithelial cells lining the lumen. The apical, luminal surface of the epithelium is protected by a dense mucus layer impenetrable to bacteria that is generated by constitutive secretion of Muc2 from luminal goblet cells. This dense layer is partially enzymatically digested to generate a looser outer layer inhabited by commensal bacteria[148]. Muc2 is secreted in a condensed conformation that expands to form a fully functional mucus layer in the presence of extracellular HCO3- and H2O. CFTR ion channel activity is required for direct export of bicarbonate and also for indirect support of export of bicarbonate and water through other channels[149]. Loss of CFTR ion channel activity results in accumulation of dehydrated mucus in the distal small intestine and proximal colon, which causes intestinal obstruction seen in CF patients in the form of meconium ileus and distal intestinal obstruction[120]. In addition, severely dilated crypts are seen in the CFTR-deficient intestine due to accumulation of mucus in goblet cells which may reflect defects in secretion[150,151]. The CFTR-deficient mucus layer appears to allow dysregulated bacterial contact. Bacterial colonies are reported in crypts with accumulated mucus[150]. Further, comparison of gene expression in the Cftr-deficient mouse intestine to changes in Muc2-deficient intestine using gene set enrichment analysis (GSEA) showed enrichment in inflammatory gene expression changes[128] suggesting that Cftr deficiency, like Muc2 deficiency, allows illicit bacterial contact. The basolateral epithelial surface and underlying lamina propria, including resident immune cells, are protected from bacterial contact by tight junctions and CFTR participates in maintenance of these junctions. In Cftr-deficient mice the small intestine displays increased intestinal permeability and disruption of tight junctions[152]. Several studies link CFTR interactions mediated by its C-terminal PDZ-binding domain to integrity of tight junctions[152]. Studies in cultured airway epithelial cells show that interaction between the CFTR-PDZ binding (CFTR PDZBD) domain and the PDZ domain of NHERF1 (SLC9A3R1) maintains actin cytoskeletal organization and tight junctions[153]. CFTR deficiency may also maintain tight junctions by via direct interaction with ZO-1 an essential component of these junctions[8].
As described above, CFTR deficiency as seen in CF patients and animal models creates an altered luminal environment. This environment, as well as a high fat/high calorie diet maintained to overcome nutritional deficits, and CF therapies such as frequent antibiotic exposure, contributes to bacterial dysbiosis[154,155]. In early studies dysbiosis was reported as small intestinal bacterial overgrowth in a CF mouse model[156]. Analysis of microbial 16S DNA in small intestine flushed luminal contents by qRT-PCR demonstrated an estimated 40X increase in bacterial load with decreased diversity[157]. In a second study using phylogenetic microarray analysis of flushed small intestinal contents there was a marked decrease in bacterial community richness, evenness and diversity, a decrease in the relative abundance of protective species such as Acinetobacter Iwoffii and Lactobacilliales members, but an increase in Mycobacteria species and Bacteriodes fragilis associated with GI infection and immunomodulation[156]. More recently, microbial population composition of CF vs non-CF individuals has been determined using targeted and metagenomic analysis of 16s rRNA DNA from fecal DNA[94,95,155,158]. Further, CF patients have been reported to be susceptible to Crohn’s Disease with specific CFTR mutations influencing microbial dysbiosis with increased intestinal permeability[159]. In each of these studies CF microbiota demonstrated greatly reduced diversity and significant differences from non-CF controls indicating that the altered luminal environment not only creates increased bacterial access but also changes bacterial composition[95,103,156]. Importantly, alterations to the intestinal microbiome are increasingly associated with CRC[103,160].
Disruption of the mucus layers and of tight junctions allows bacterial access to immune cells and immune infiltration of the epithelial layer. Although immune infiltration and subsequent inflammation can cause overt damage, this effect has only been reported in one study in which a capsule endoscopy study of CF patients revealed duodenal lesions[161]. More commonly, immune infiltration has been detected under CFTR-deficient conditions but has not been accompanied by obvious morphological and histological damage[162]. In human studies CF patients exhibited evidence of immune infiltration in studies evaluating whole gut lavage[163], fecal calprotectin[158,161,164] and rectal nitric oxide[165]. Further, microarray analysis of the small intestine in an early study by Norkina et al[162] identified upregulation of genes expressed by granulocytes which was supported by microscopy that identified increased mast cells and neutrophils throughout the length of the intestine. Similarly, Chan and colleagues reported immunocytochemical evidence of an increased presence of macrophage and neutrophils in the small intestine of F508del mice[145].
Loss of physical barriers in the GI tract allows dysregulated access of bacteria and infiltration of immune cells both of which contribute to inflammatory signaling[166-169]. These contacts have the potential to activate pro-inflammatory signaling in the intestinal epithelium both directly by interaction between bacteria and epithelial toll-like receptors[170] and indirectly via activation of lymphocyte cytokine signaling. For example, Fiorotto et al[171] demonstrated that in primary biliary epithelial cells CFTR binds Src inhibitors to position them for interaction with Src. Loss of CFTR leads to mislocalization of these inhibitors and activation of Src. In the presence of bacterial products this leads to NF-κB signaling which ultimately disrupts tight and adherens junctions. Proinflammatory signaling by intestinal epithelial cells is associated with dysregulated proliferation and expansion of the stem cell compartment through reversion of progenitor cells to stem cells[172]. Loss of barrier integrity affects other processes as well. Loss of tight junctions facilitates increased migration and invasion[173]. Dysbiosis may result in appearance of bacterial species associated with CRC. Phylogenetic microarray analysis of Cftr-deficient mice detected increased representation of Bacteroides fragilis[156], a species directly linked to CRC by virtue of its activation of Stat3 signaling through a Th17 immune response[174].
CFTR deficiency has been linked to NF-κB activation in in vitro models. Chan and colleagues have carried out a series of studies in cell lines derived from several cancers, that delineate a pathway in which CFTR deficiency leads to activation of NF-κB, transcription of UPA, and enhanced migration and invasion[175-177]. Other groups have shown increased basal levels of pro-inflammatory cytokines and other factors and NF-κB pathway members in CFTR-deficient Caco-2, HT-29 and HEK293 cells and increased response to inflammatory stimuli, including an increase in TNF-α, IL-6, IL-1β-induced secretion of IL-8, COX-2 and PGE2, plus increased activities of ERK1/2, MAPK, IκBα and NF-κB[92,96,178]. Santa-Coloma and colleagues report activation of NF-κB as a result of autocrine signaling by IL-1β[179]. In addition, the Hedgehog pathway has recently been reported to be inhibited under CFTR-deficient conditions which is predicted to activate inflammatory signaling[180].
CFTR deficiency has been reported to both increase oxidative stress via impaired mitochondrial activity[96,97] and to reduce stress by cellular retention of the antioxidant GSH[181]. In Caco-2/15 cells CFTR knockdown caused an increase in lipid peroxidation levels accompanied by a decrease in oxidant defenses such as glutathione peroxidase and catalase[182]. In addition, loss of CFTR has been reported to disable autophagy via activation of Transglutaminase-2 whose cross-linking activity causes essential autophagy proteins to be sequestered in aggresomes[183]. Finally, HIF-1-mediated repression of CFTR in intestinal epithelium resulted in a reduction in CFTR mRNA, protein and activity in hypoxic epithelium[184]. The impact of these activities on oncogenesis is not yet known.
CFTR deficiency results in dysregulation of many processes that may be oncogenic. However, the relative contributions of these processes to the development of CRC in vivo are not yet clear. In the future it will be important to study CFTR in the environment in which it most likely contributes to development of CRC, the intestinal stem cell compartment. In the colon, CFTR is most highly expressed in the base of the crypt which comprises the stem cell compartment and the likely site of origin of CRC. Because the crypt is the environment for the intestinal stem cell it has unique protective mechanisms. The crypt maintains a unique microbiota with reduced number and diversity compared to the lumen[185]. Limited contact between bacterial products and crypt epithelial cells contributes to this protected environment. For example, goblet cells at the neck of the crypt orchestrate the release of mucin in response to contact with bacterial products[186]. The crypt also has unique mechanisms to protect from cellular stress. In the stem cell, interaction between the cytosolic innate immune sensor NOD2 and the bacterial peptidoglycan motif MMP is necessary for stem cell survival in the face of oxidative stress[187]. Finally, cells in this compartment are uniquely susceptible to the effects of inflammatory signaling. In a mouse genetic study NF-κB was shown to synergize with Wnt/β-catenin in intestinal crypt cells to promote conversion of progenitor cells to stem-like cells with tumor initiating capacity[172]. These considerations highlight the importance of studying potential oncogenic phenotypes of CFTR deficiency in the crypt and crypt-derived organoid models.
Given the widespread distribution of CFTR and its impact on cellular homeostasis it is not surprising that dysregulation of CFTR is implicated in many cancers. CFTR overexpression has been correlated with cancer in individual studies, in particular, gastric and ovarian cancers[101,188-192]. However, in the vast majority of studies CFTR deficiency is associated with cancer occurrence, and in most of these studies, cancers with CFTR mutations or down-regulation of expression in tumors are more likely to exhibit rapid expansion, EMT, decreased apoptosis, increased metabolic potential, and increased patient morbidity and mortality. Decreased CFTR expression and/or inactivating mutations are associated with non-small cell lung cancer[130,175,193], glioblastoma[194], bladder[131-133], EC[195,196], PC[197,198], nasopharyngeal[199], prostate[176], and breast[177] cancers. In most of these cancers CFTR deficiency was associated with increased tumor stage and poor survival. Germline CFTR mutations, as seen in CF, have been linked to increased risk of PC among younger patients[197,198]. Germline mutations may be an important risk factor in the general population as an estimated 3% of the United States population are heterozygote carriers for deleterious CF-causing CFTR mutations[200] and therefore potentially at risk due to loss of heterozygosity or haploinsufficiency. However, in most studies CFTR deficiency was associated with differentially decreased expression but not specifically with germline defects. Although germline mutations were not examined in these cases it is unlikely that decreased expression was caused primarily by CF-causing germline mutations because F508del, which makes up approximately 70% of CF alleles, causes a 90% decrease in protein levels but only a modest decrease in mRNA. Known causes of decreased CFTR expression include hypermethylation as seen in lung and bladder cancer studies[130-133] and somatic mutations seen in a NSCLC study[130]. In addition, cigarette smoke (CS) has recently been shown to down-regulate CFTR expression and CFTR down-regulation has been linked to the etiology of COPD[201]. However, a recent study also linked CS-mediated down-regulation of CFTR to exacerbation of oncogenic phenotypes in the A549 lung cancer cell line[202].
In the era of precision medicine new CF modulator therapies targeted to specific CFTR mutations have entered the clinic. These include potentiator drugs that increase anion flow through CFTR channels already present on the PM and corrector drugs that promote correct folding of mutant proteins. Three modulators, ivacaftor, tezacaftor, and lumacaftor, have been approved for treatment of CF[203]. Ivacaftor is a potentiator approved for treatment of > 25 CF-causing mutations including gating, residual function and splice mutations. Tezacaftor and lumacaftor are correctors designed to improve the function of the F508delta mutation which makes up approximately 70% of CF alleles. As discussed by Bodewes et al[204] these drugs are finally targeting CF-related GI diseases, with potential use in cancer therapy. Some examples include treatment of pancreatitis with Ivacaftor[204], drug treatment of CF patients that improved proximal small intestine pH as a regulator of bicarbonate secretion, improvement in cell motility and clinical outcomes in patients with CFTR G551D mutations[104], improvement in bicarbonate permeability following Lumacaftor-rescued F508del mutations[205], improvement in gut microbiota and intestinal inflammation following treatment with Ivacaftor, including an increase in Akkermansia and a decrease in Enterobacteriaceae, and a significant reduction in inflammation in patients treated with Ivacaftor[206]. Additional modulators and combinations of modulators are under development such as the combination of Ivacaftor with 5-Nitro-2-(3-Phenylpropylamino) Benzoate, that is reported to act synergistically in activation of G551D mutant CFTR[207]. These mutation-specific drugs are potentially applicable to the 3% of Caucasians who are CF carriers, having one CF-causing CFTR mutation. These individuals are potentially at risk for developing CRC due to haploinsufficiency or loss of heterozygosity. In addition to these Food and Drug Administration (FDA)-approved therapies, another category of modulator known as amplifiers, represented by PTI-428, is in clinical trials. This drug is designed to enhance translation of CFTR mRNA to increase protein production and facilitate the activity of corrector drugs[99]. Thus, this drug has the potential to improve CFTR activity in sporadic CFTR-low CRC. In addition, testing strategies used for screening and diagnosis of CF may be applicable to early detection and even prevention of CRC. Currently genetic testing for CF-causing mutations is recommended for all pregnant couples. If carrier status proves to be a risk factor for CRC, then this genetic testing may be adaptable for early detection of CRC as well. A significant technical advance to test new CF therapies has been the development of patient-derived colorectal organoids. An example is the work of Dekkers et al[208] who have demonstrated proof of this idea in rectal organoids derived from CF patients to test a range of CF drugs, including Ivacaftor and Lumacaftor. While these new targeted CF therapies have yet to enter the cancer clinic, they may soon do so, along with the repurposing of other ion channel activators and blockers as more becomes known about the precise contribution of ion channels to cancer development.
Ca2+ activated Cl- channels (CLCAs) are a family of secreted self-cleaving proteins that activate Ca2+-dependent Cl- channels. CLCAs are involved in Cl- conductance across epithelial cells and therefore influence epithelial fluid secretion, cell-cell adhesion, apoptosis, cell cycle control, tumorigenesis and metastasis, mucus production and cell signaling[209,210]. There are four CLCAs (1-4) in humans and two of these family members (CLCA1,2) are implicated in GI cancers, almost invariably as tumor suppressors whose down-regulation is associated with cancer progression and poor patient prognosis. CLCA1 is down-regulated in CRC and this is associated with poor patient prognosis[4,211,212]. CLCA1 has also been reported to suppress CRC aggressiveness via inhibition of the Wnt/β-catenin signaling pathway[213]. CLCA1 is also reported to be a prognostic biomarker for PC, with lower CLCA1 expression correlated with shorter disease-free survival[214]. CLCA2 is also down-regulated in CRC[4,212].
Cl- intracellular channels (CLICs) are novel, auto-inserting, self-assembling intracellular anion channels involved in a wide variety of fundamental cellular events including regulated secretion, cell adhesion, cell cycle and apoptosis. However, the actions of CLICs remain to be fully explained. They are a class of intracellular anion channels that do not resemble classical ion channel proteins. CLICs can exist as both soluble globular proteins and integral membrane proteins with ion channel function. Human CLIC1 adopts at least two stable soluble structures, with redox status controlling the transition between them. CLIC proteins are characterized by the presence of a 240 residue CLIC module whose structure belongs to the glutathione S-transferase fold superfamily[215]. Three CLICs appear to be functional in humans (CLIC1,3,4) with two CLIC proteins (CLIC1 and CLIC4) that appear to be essential molecular components of anion channels, with CLIC1 capable of forming anion channels in planar lipid bilayers in the absence of other cellular proteins. However, these putative ion channel proteins are controversial because they exist in both soluble and membrane forms, with at least one transmembrane domain[209,215]. All three CLICs are involved in GI cancers, with all implicated as oncogenes. CLIC1 is over-expressed in CRC[216-219]; in PC[65,217], where it was reported to be upregulated in 69% of tumors and associated with poor patient prognosis; and in GC[65,218,220-223], where it was found upregulated in 68% of tumors, correlated with lymph node metastasis, lymphatic invasion, perineural invasion and poor patient prognosis. It was also reported to promote GC progression by regulating ROS-mediated MAPK/AKT signaling. CLIC1 was also reported to be upregulated in HCC[65,224] and gall bladder cancer[225,226]. CLIC3 is upregulated in PC where it was reported to promote integrin recycling from late endosomes to drive PC progression[65,227]. CLIC3 is also a secreted protein that is reported to drive cancer progression through its glutathione-dependent oxireductase activity. In particular CLIC3 was identified as part of the cancer associated-fibroblast secretome where it promotes the invasive behavior of endothelial cells to promote angiogenesis and invasion. CLIC3 is also secreted by cancer cells[228], and CLIC3 is described as a pH sensor, important as changes in cellular pH influence cell proliferation and the balance between cell survival and cell death[229]. CLIC4 is upregulated in CRC where it was found to be a direct response gene for C-MYC and TP53, with its overexpression associated with poor 5-year patient survival[218,230]. CLIC4 is also upregulated in PC with its expression associated with tumor grade, invasion and poor patient survival[231]. CLIC4 was also found to be expressed in mitochondria where it regulates pH and cell volume.
ANO1 is also referred to as Anoctamin-1, DOG1 and TMEM16A. ANO1 is a Ca2+-activated Cl- channel that mediates receptor-activated Cl- currents that are involved in a range of physiological processes. ANO1 is activated by intracellular Ca2+. ANO1 has 8 putative transmembrane domains and demonstrates no similarity to other ion channels. It is expressed in a variety of secretory epithelia, including the gut. ANO1 activity is implicated in regulating CFTR Cl- channel activity. In all of the GI cancers studied, ANO1 expression and activity has been reported to upregulated, thus ANO1 can be described as an oncogene in the GI tract. In CRC, ANO1 expression is upregulated, associated with EMT and poor patient prognosis[232-236]. In PC, ANO1 expression is important for cell migration[234,237]. In EC, ANO1 is a biomarker of EC progression[234,238]. In GC, its expression is upregulated[234,239], and in GI stromal tumors, ANO1 expression is used as a diagnostic biomarker, and it is associated with negative regulation of IGFBP5[240-242]. See Table 2 for a full listing of chloride ion channels in GI cancers.
| Gene name (s) | Cancer | Role | Functional activity |
| CLCA1/Chloride channel accessory 1 | Colorectal | Tumor suppressor | Expression down-regulated; down regulated in primary tumors and CRC cells; acts via inhibition of Wnt/β-catenin signaling; there is one report that high expression associated with non-response to chemo radiation therapy in rectal cancer[4,211-213] |
| Pancreatic | Tumor suppressor | Expression down-regulated; low expression associated with poor patient prognosis[214] | |
| CLCA2/Chloride channel accessory 2 | Colorectal | Tumor suppressor | Expression down-regulated[4,212] |
| CLIC1/Chloride intracellular channel 1 | Colorectal | Oncogene | Expression up-regulated; overexpressed by MS analysis of human CRCs; expressed on nuclear and plasma membranes[216-219] |
| Pancreatic | Oncogene | Expression up-regulated; over expression associated with poor patient prognosis, tumor grade and size; overexpression in 69% of tumors; knockdown of PC cells reduced cell proliferation and anchorage-independent growth on soft agar, and cell migration[65,217-219] | |
| Gastric | Oncogene | Expression up-regulated; overexpression associated with poor patient prognosis; upregulated in 68% of gastric cancer, correlates with lymph node metastasis, lymphatic invasion, perineural invasion and pathological staging; induced proliferation, apoptosis, invasion and migration of GC cells in culture; promotes progression by regulating MAPK/AKT pathway; regulates migration and invasion via ROS-mediated P38 MAPK pathway[65,220-223] | |
| Hepatocellular | Oncogene | Expression up-regulated[65,224] | |
| Gall bladder | Oncogene | Expression up-regulated; knockdown in GBC cells reduced proliferation, migration and invasion of cells; associated with metastasis, based on proteomic analysis[284-286] | |
| CLIC3/Chloride intracellular channel 3 | Pancreatic | Oncogene | Expression up-regulated; CLIC3 and Rab25 collaborate to promote integrin recycling from late endosomes/lysosomes to drive PaC progression[65,227] |
| CLIC4/Chloride intracellular 4 | Colorectal | Oncogene | Expression up-regulated; associated with poor 5-yr patient survival; CLIC4 regulated by TP53 and TNF-alpha and is a direct response gene for C-MYC and TP53[218,230] |
| Pancreatic | Oncogene | Expression up-regulated; associated with tumor grade, lymph node metastasis, tumor invasion and poor patient survival; expressed in mitochondria and regulates pH and cell volume[231] | |
| CFTR/Cystic fibrosis transmembrane conductance regulator | Colorectal | Tumor suppressor | Expression down-regulated; CF patients at significant risk of early aggressive colorectal tumors based on colonoscopy screening and other clinical findings; CFTR down-regulated in sporadic CRC, associated with worse prognosis; CFTR was a CIS gene in four sleeping beauty transposon mutagenesis screens in mice, both CRC and GC; > 60% conditional CFTR KO mice develop intestinal tumors and crossing to ApcMin model causes an enhanced tumor phenotype and the development of adenocarcinomas; enhanced organoid outgrowth; CFTR deficiency causes an invasive phenotype in CRC cells; loss of CFTR causes upregulation of NF-κB; CFTR modulates Wnt/β-catenin signaling and stem cell proliferation; enhanced risk of CRC in CF patients following lung trans-plants[33-34,36,93,103,119,123-126,128,243-247] |
| Pancreatic | Tumor suppressor | Expression down-regulated; increased risk of PC in carriers of 4 specific CFTR mutations[101,118,122,123,197,198,248-250] | |
| Small intestine | Tumor suppressor | Expression down-regulated[122-123,243] | |
| Gastric | Oncogene | Expression up-regulated in late stage[101,191,192] | |
| Esophageal | Tumor suppressor | Expression down-regulated; silencing of CFTR caused upregulation of NFKB; CFTR inhibitors caused enhanced growth of EC cell mouse xenografts; enhanced risk of EC in CF patients following lung transplants; CFTR heterozygous carriers at enhanced risk of EC[195,196,243,251] | |
| Hepatocellular | Tumor suppressor | Expression down-regulated by promoter hypermethylation[89,252] | |
| ANO1/anoctamin1/TMEM16A/DOG1 | Colorectal | Oncogene | Expression up-regulated; negatively regulated by miR-144 miR-9, and miR-132; associated with EMT and poor patient prognosis[232-236] |
| Pancreatic | Oncogene | Expression up-regulated; important for cell migration[234,237] | |
| Esophageal | Oncogene | Expression up-regulated; biomarker for EC progression[234,238] | |
| Gastric | Oncogene | Expression up-regulated[234,239] | |
| GI stromal (GIST) | Oncogene | Expression up-regulated; used as a diagnostic biomarker; associated with negative regulation of IGFBP5[240-242] |
Ca+ is a ubiquitous second messenger, serving as a signaling molecule for a variety of cellular processes such as control of the cell cycle, apoptosis, and migration. Ca+ concentrations are tightly regulated within the cell, with basal cytoplasmic levels being many orders of magnitude less than in the extracellular space. This control is essential for cellular homeostasis[253]. In addition to extracellular Ca2+, the endoplasmic reticulum (ER) and mitochondria are also significant stores of Ca2+. Selective distribution and activation of Ca2+ channels at any of these sources allows for Ca2+ micro-domains and adds another level of specificity to Ca2+ signaling[6]. Because it is involved in such a broad array of processes, and signaling molecules are often sensitive to very minute changes in Ca2+, it is easy to see how perturbations in Ca2+ handling could lead to significant physiological outcomes.
Ca2+ signaling is typically initiated through the ligand activation of various receptors that activate phospholipase C[254]. This leads to the production of inositol triphosphate, which diffuses to the ER, where it binds to and opens its receptor and Ca2+ channel. Upon its release into the intracellular space, Ca2+ binds to calmodulin and a variety of other Ca2+-activated proteins to elicit a wide variety of responses. Over time, these intracellular stores would be depleted if not for being replenished by extracellular Ca2+. The release and depletion of ER Ca2+ triggers the process of store-operated calcium entry (SOCE), in which PM Ca2+ channels are opened in order to allow entry of extracellular Ca2+[254]. One of the proteins responsible for sensing the depletion of ER stores, and initiating SOCE, is stromal interaction protein 1 (STIM1)[254]. In CRC, STIM1 over-expression has been associated with increased tumor size, depth of invasion, lymph node metastasis, and increased serum levels of carcinoembryonic antigen[15,255,256]. When injected into immunocompromised mice, CRC cells expressing a higher level of STIM1 had a much higher incidence of lung and liver metastasis than those expressing lower levels[257].
The PM Ca2+ channels opened by STIM1 activation include ORAI1 as well as members of the TRP superfamily of cation channels. In addition to replenishing intracellular stores, these PM channels also contribute to Ca2+ signaling in response to intracellular or extracellular cues. Different members of the TRP superfamily are activated by physical changes such as mechanical stress, osmotic pressure, or temperature; or chemical changes such as pH, growth factors/cytokines, pO2, or ROS[258]. With all of these being central factors in a tumor microenvironment, how a cell alters its response to these conditions could ultimately influence the death or survival of a potentially cancerous cell.
Various members of the transient receptor potential (TRP) superfamily have been implicated in GI cancers, in particular the TRPM (m = melastatin) and TRPC (c = canonical) sub-families. The cold-activated TRPM8, well known for its role in androgen-dependent prostate carcinoma, is also over-expressed and necessary for the proliferation and migration of PC cells[259-262]. TRPM7 also plays a role in PC, but by increasing motility and thus the potential for metastasis, with its depletion causing a decrease in invasiveness through the HSP90α/uPA/MMP-2 proteolytic axis and targeted senescence, while results vary as to its role in proliferation[5,258-261,263-267]. TRPM7 has also been implicated as an oncogene in CRC[268] and GC[269-271]. Though a specific mechanism has not been proposed, TRPM2 over-expression has also been shown to reduce PC as well as GC patient survival by increasing invasiveness and pro-liferation[272,273]. Expression of TRPC1 is reported to be upregulated in CRC[5,256,274], where it promotes metastasis, EC[275], and GC[68]. TRPC6 expression is upregulated in EC[276,277] where it was found to be necessary for Ca2+ increase to promote G2 progression, and association with tumor stage and poor patient prognosis, GC[5,278], and HCC[4].
Through its contribution to hyperactivity of intracellular Ca2+ oscillations, ORAI1-high cells had increased activation of ERK, AKT, and myocyte enhancer factor 2D, indicating a possible mechanism explaining their increased proliferative and invasive phenotype[275]. ORAI1 expression is also upregulated in CRC[5], where it is activated by STIM1; PC[92,279], where it mediates SOCE and promotes apoptotic resistance in PC cells; GC[68], where it is associated with promoting metastasis; and in EC, where ORAI1 over-expression had a negative effect on patient prognosis.
Other oncogenic Ca2+ channels include SIGMAR1 in CRC[280-282], L-TYPE CACNA1C in CRC[283], T-type CACNA1I in GC[284], where its upregulation is associated with poor patient survival, and T-type CACNA1H in GC[284], where it is also associated with poor patient survival.
While most Ca2+ channels appear to function in an oncogenic fashion when dysregulated there are several exceptions. TRPM6 has been reported to be down-regulated in CRC (16 of 20 tumors studied) with high expression associated with better patient survival[285]. STIM2 is reported to be down--regulated in CRC leading to apoptosis resistance[15,256,274]. Expression of the T-type channel CACNA1G is reported to be down-regulated by promoter hypermethylation in CRC[286], PAC[287], and GC[288], with high expression associated with improved overall survival. CACNA2D3 expression is down-regulated by promoter hypermethylation, associated with worse patient prognosis[91,289] and expression of CACNB2 is also reported to be down-regulated by promoter hypermethylation[90]. See Table 3 for a full listing of the role of calcium ion channels in GI cancers.
| Gene name (s) | Cancer | Role | Functional activity |
| TRPC1 | Colorectal | Oncogene | Expression up-regulated; promotes metastasis[5,256,274] |
| Esophageal | Oncogene | Expression up-regulated[275] | |
| Gastric | Oncogene | Expression up-regulated[276] | |
| Hepatocellular | Oncogene | Expression up-regulated[249] | |
| TRPC6 | Esophageal | Oncogene | Expression up-regulated; necessary for Ca2+ increase to promote G2 progression; associated with tumor stage and poor prognosis[276,277] |
| Gastric | Oncogene | Expression up-regulated[5,278] | |
| Hepatocellular | Oncogene | Expression up-regulated[4] | |
| TRPM2 | Colorectal | Oncogene | Expression up-regulated[270] |
| Pancreatic | Oncogene | Expression up-regulated; enhanced proliferation, invasion & metastasis[272,273] | |
| Gastric | Oncogene | Expression up-regulated; inhibition reduced proliferation of gastric cancer cells, increased autophagy and sensitized cells to paxlitaxel and doxorubicin[68,272,273] | |
| TRPM6 | Colorectal | Tumor suppressor | Expression down-regulated in 16/20 (80%) of primary tumors; high expression associated with better patient survival[285] |
| TRPM7 | Colorectal | Not determined | Genetic polymorphism associated with enhanced risk of adenomas, linked to high Ca2+:Mg2+ ratio in diet[268] |
| Pancreatic | Oncogene | Expression up-regulated; increased tumor growth, invasiveness and metastasis; targeted silencing induced replicative senescence[5,258-261,263-267] | |
| Gastric | Oncogene | Highly expressed in gastric cancer cell lines; required for GC survival linked to Mg; suppression induces cell death in culture[4,269-271] | |
| TRPM8 | Pancreatic | Oncogene | Expression up-regulated; regulates proliferation and migration; silencing in cell lines induces replicative senescence [259-262] |
| L-type/a1c subunit/CACNA1C | Colorectal | Oncogene | Expression up-regulated[283] |
| Sig1R/SIGMAR1 | Colorectal | Oncogene | Expression up-regulated in CRC cell lines and primary CRC tumors[280-282] |
| Stim1/Stromal interaction protein 1 | Colorectal | Oncogene | Expression up-regulated; increased CRC cell motility; STIM1 overexpression enhanced lung and liver metastases in mouse xenograft models; also associated with poor prognosis in CRC patients[15,255,256] |
| Pancreatic | Oncogene | Expression up-regulated; promotes invasion and metastasis; STIM1 and Orai1 are the molecular components of SOCE[14] | |
| Stim2/Stromal interaction protein 2 | Colorectal | Tumor suppressor | Expression down-regulated; depletion causes apoptosis resistance[15,256,274] |
| Orai1/CRAMC1 | Colorectal | Oncogene | Expression up-regulated; activated by STIM1[5] |
| Pancreatic | Oncogene | Expression up-regulated; mediate SOCE and promote apoptotic resistance in pancreatic cancer cells[91,279] | |
| Esophageal | Oncogene | Expression up-regulated; promotes tumor-promoting Ca2+ oscillations in EC[275] | |
| Gastric | Oncogene | Expression up-regulated; promotes metastasis[68] | |
| T-type CACNA1G/CaV3.1 | Colorectal | Tumor suppressor | expression down-regulated by promoter hypermethylation[284,286] |
| Pancreatic | Tumor suppressor | Expression down-regulated by promoter hypermethylation[284,287] | |
| Gastric | Tumor suppressor | Expression down-regulated by promoter hypermethylation; high expression associated with improved overall survival[284,288] | |
| T-type CACNA1I/CaV3.3 | Gastric | Oncogene | High expression associated with poor survival[284] |
| T-type CACNA1H/CaV3.2 | Gastric | Oncogene | High expression associated with poor survival[284] |
| CACNA2D3 | Gastric | Tumor suppressor | Expression down-regulated by promoter hypermethylation, associated with worse prognosis[91,289] |
| CACNB2 | Esophageal | Tumor suppressor | Expression down-regulated by promoter hypermethylation[90] |
Voltage-gated sodium channels (VGSCs) are classically associated with the initiation and conduction of action potentials in electrically excitable cells such as neurons and muscle cells. They are typically heteromeric complexes composed of one of 9 pore-forming α-subunits and one of 5 regulatory β-subunits, though the α-subunit can usually be a functional channel by itself. These channels have recently been discovered in non-excitable cell types such as glia, fibroblasts, immune cells, and cancer cells, where their function is less understood[290]. In the developing central nervous system, VGSCs regulate the migration and pathfinding of neurite outgrowth[291-293]. Similarly, in the vast majority of cancers where these channels have been studied, their major influence has been to increase the motility and invasiveness of cancer cells.
The VGSC NaV1.5 is abundantly expressed in human CRC cells SW620, SW480, and HT29, as well as being highly expressed in tumor biopsies relative to adjacent normal tissue[294]. Through pharmacological inhibition, siRNA knockdown, and controlling for Ca2+ influence, House et al[294] demonstrated that conductance through NaV1.5 contributes significantly to CRC cell invasiveness and cancer progression. This group later demonstrated this contribution mechanistically through correlating NaV1.5 activity to RAP-1 mediated MAPK activation and changes in gene expression through the PKA/ERK/c-Jun/ELK-1/ETS-1 signaling pathway[295]. While this group demonstrated the metastatic contribution of NaV1.5, Peng et al[296] have recently shown Nav1.5 expression to be predictive of clinical outcomes in non-metastatic CRC, with the potential involvement of estrogen receptor (ER)-β, implicating multiple roles for the same channel in different stages of cancer progression.
In GC, high expression of the VGSC Nav1.7 has similarly been shown to correlate with higher recurrence and reduced survival[297]. The detailed mechanism proposed by Xia et al[297] involves increased expression of the oncoprotein metastasis-associated in colon cancer-1 (MACC1) by Nav1.7 activation in a p38/NFkB-dependent manner. Increased MACC1 expression subsequently leads to HGF/c-Met/c-Jun-dependent activation of another oncoprotein, the Na+/H+ exchanger-1 (NHE1) which, through its involvement in maintaining intracellular and extracellular pH, has been shown elevated in a variety of cancers[298-301]. Reports involving other Nav channels in GI cancer are fewer than for Nav1.5 and Nav1.7. Nav1.1 has been reported to be upregulated in CRC[302], associated with a shorter time to cancer recurrence in stage II and III disease. In contrast, there is one report of Nav1.6 being down-regulated in CRC[303].
The mechanisms through which VGSC α and β subunits contribute to cancer progression typically differs according to their physiological functions: α subunits, through the conduction of Na+ currents, and β subunits through the regulation of adhesion interactions, though contributions of any β subunit have yet to be shown in a GI cancer. The reason that only α subunits have been associated with GI cancers may owe in part to these differences in function relative to the demands of GI epithelial physiology. This specificity may also prove advantageous in pharmacologically targeting these subsets of cancers, as drugs targeting α subunit function have proven effective in reducing metastasis in murine xenograft breast cancer models[304,305]. See Table 4 for a full listing of sodium ion channels in GI cancer.
| Gene name (s) | Cancer | Role | Functional activity |
| SCN1A/Nav1.1 | Colorectal | Oncogene | Time to reoccurrence of stage II and III CRC is shorter in patients carrying Nav1.1 variants[302] |
| SCN5A/Nav1.5 | Colorectal | Oncogene | Expression up-regulated; mediates invasion via MAPK signaling; key regulator of a transcriptional network that includes Wnt/β-catenin signaling; associated with poor patient prognosis; linked to upregulation of ER-β[5,294-296] |
| SCN8A/Nav1.6 | Colorectal | Tumor suppressor | Expression down-regulated in CRC tumor tissues compared with control[303] |
| SCN9A/Nav1.7 | Gastric | Oncogene | Expression up-regulated; mechanistically related to upregulation of MACC1 and NHE1[297] |
Zinc is a key trace element in the human body. Approximately 10% of human genes have the potential for zinc binding and a large number of human proteins (as many as 3000), including transcription factors, a variety of receptors, kinases, ligases and other enzymes, require zinc for their catalytic activity or tertiary structure[306,307]. Zinc exists in cells as Zn2+ ions that are maintained in this valence state under all biologically relevant redox potentials and pH conditions in a cell. Zinc mediates a wide range of cellular processes that are important for maintenance of tissue homeostasis[306,308,309]. Therefore, alterations in cellular zinc levels, such as zinc deficiency, can disrupt cellular function[306,308]. Furthermore, zinc is toxic to cells, thus zinc levels are tightly regulated within cells. As zinc cannot freely pass across cellular membranes a variety of Zn2+-permeable channels and transporters regulate Zn2+ entry and exit from cells[306]. Zinc transporters include both influx and efflux transporters[306,310,311]. Zinc also enters cells via other ion channels, such as Zn2+ activation of several of the Zn2+-permeable voltage-gated Ca2+ ion channels[309]. Examples include the store-operated Ca2+ entry (SOCE) channels and the TRP family of Ca2+ channels (discussed earlier in this review).
Here, the review will focus on the functions of the two major families of Zn2+ transporters: ZnT/SLC30A and ZIP/SLC39A. Members of both of these families of proteins are reported to be dysregulated in human cancers, including GI cancers. There are 14 ZIP proteins (ZIP1-ZIP14) and 10 ZnT proteins (ZnT1-ZnT10) in the human body with differential tissue-specific expression[310,311].
These proteins are encoded by the solute carrier family 39A genes (SLC39A1-SLC39A14). ZIPs mainly participate in the uptake of Zn into the cytoplasm from the extracellular space or from intracellular compartments such as the ER, Golgi or mitochondria. In the GI tract ZIPs 1, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 14 are reported to be normally expressed in the intestines; ZIPs 1, 3, 4, 5, 6, 7, 8, 9, 10, and 14 in the pancreas; ZIPs 1, 2, 3, 5, 6, 7, 8, 9, and 14 in the liver; and ZIPs 1, 3, 4, 5, 6, 7, 8, 9 10, and 11 in stomach[310,312,313].
These proteins are encoded by the solute carrier family 30A genes (SLC30A1- SLC30A10). ZnTs participate in the efflux of Zn from the cytoplasm into the extracellular space or to sequester Zn in intracellular compartments. In the GI tract, ZnT1 expression is normally expressed ubiquitously; ZnTs 2, 3, 5, 6, 7 and 10 are normally expressed in the intestines; ZnTs 2, 3, 5, 6, 7 and 8 in pancreas; and ZnT 5, 6, 7 and 10 in liver and ZnT 6, and 7 in stomach[310,312,313].
It is well known that dysregulation of zinc homeostasis results in GI dys-function[311,312,314,315]. Examples include zinc deficiency resulting in diarrhea[311], and inflammatory bowel disease[316], clinical problems that are ameliorated by zinc supplementation[311,316,317]. This effect of zinc supplementation may be mediated by a zinc sensing receptor molecule, GPR39, that is similarly shown to be protective in the colon[318]. Zinc is also reported to enhance tight junctions and intestinal mucosal barrier functions in general[319,320]. Maintenance of zinc homeostasis is primarily mediated by zinc transporters that act to absorb diet-derived zinc for distribution to the peripheral tissues[321]. But excess zinc is toxic to both intestinal epithelial cells[308,315] and also peripheral tissue cells, thus zinc transporters maintain cytosolic zinc levels via both influx and efflux of zinc ions[306]. The physiological role of zinc and zinc transporters have been best studied in the intestinal tract, where zinc is required for intestinal epithelial homeostasis, a process mediated by several zinc transporters. In intestinal absorptive enterocytes, ZIP4 and ZnT1 regulate zinc absorption, while ZIP5 and ZnT5B regulate zinc excretion. In intestinal Paneth cells, ZnT2 and ZIP4 are required for zinc accumulation in Paneth cell granules and are important for Paneth cell maintenance overall. Of potential importance to intestinal cancer, ZIP4 and ZIP7 are expressed in the intestinal crypt stem cell compartment where they contribute to Lgr5+ stem cell maintenance and help maintain transit amplifying cell proliferation[312].
The role of zinc transporters in human cancer have been best studied in prostate and breast cancer[308,322]. In the prostate gland ZIPs, especially ZIP1, are considered tumor suppressors[323]. In breast cancer, increased levels of zinc correspond to upregulation of several ZIPs (and a few ZnTs as well), thus zinc transporters as a group are considered as oncogenes in breast cancer[324]. In the GI tract, the majority of reports show upregulation of both ZIPs and ZnTs in all major GI organ cancers (Table 5), although for some cancers there is clearly conflicting evidence. The strongest evidence for dysregulation of zinc transporters in GI tract cancers has been for PC, where zinc transporter upregulation has been associated with enhanced cancer cell migration and worse patient prognosis. This is especially true for ZIP4. In one study increased mRNA expression of ZIP4 was observed in 16 of 17 pancreatic adenocarcinoma samples[325], a finding that was supported in ZIP4-expressing xenografts in mice that yielded larger tumors[326]. These findings were confirmed by several other groups[327-329], such as Xu et al[329] who found that ZIP4 was upregulated in a set of 23 PCs and that ZIP4 expression could be used as a diagnostic and prognostic marker. While the case of ZIP4 as an oncogene is very clear, for other zinc transporters the data is sometimes contradictory, with some reports of upregulation and others of down-regulation of non-ZIP4 zinc transporters[313,330]. Studies employing zinc-sensitive histochemical staining have reported a general reduction in zinc levels in PC samples[331,332], a report that is difficult to reconcile with upregulation of zinc transporters, although it has also been noted that increases in zinc transporter mRNAs do not always correspond to increased protein expression and activity as many zinc transporters, including ZIP4, are regulated by posttranslational mechanisms[322]. Further, some studies have shown down-regulation of virtually all zinc transporters in PC, with the exception of ZIP4[313]. Another case for zinc transporters acting as an oncogene or tumor suppressor is in EC, where an inherited genetic variant in the 5’ untranslated region of ZIP6 results in constitutive expression of ZIP6, enhancing the malignancy of ESCC cells[333]. In ESCC cell lines upregulation of ZIP6 enhances proliferation, migration, and invasion of cancer cells, while down-regulation of ZIP6 suppresses these effects[333]. Similarly, Jin et al[334] reported that knockdown of ZIP5 significantly inhibited human ESCC cell progression and Kumar et al[335] reported upregulation of ZIP7 in ESCC. In contrast, other studies have reported that zinc deficiency contributes to progression of EC. Choi et al[336] reported that zinc supplementation inhibited proliferation of ESCC cell lines, a process mediated by inhibition of Orai-mediated SOCE and subsequent Ca2+ oscillations. For other GI cancers such as CRC and GC, there is less evidence but those reports point to an oncogenic role for zinc transporters[337-339], despite zinc’s known protective effects against colonic inflammation.
| Gene name | Cancer | Role | Functional activity |
| ZnT1/SLC30A1 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZnT2/SLC30A2 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZnT3/SLC30A3 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZnT4/SLC30A4 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZnT5/SLC30A5 | Colorectal | Oncogene | Increased mRNA expression in tumors[337] |
| ZnT6/SLC30A6 | Colorectal | Oncogene | Increased mRNA expression in tumors[337] |
| ZnT7/SLC30A7 | Esophageal colorectal | Oncogene oncogene | Increased mRNA expression in tumors[308,335] Increased mRNA expression in tumors[337] |
| ZnT8/SLC30A8 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZnT9/SLC30A9 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| ZIP1/SLC39A1 | Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] |
| Pancreatic | Tumor suppressor | Down regulated mRNA expression in tumors[313] | |
| ZIP2/SLC39A2 | Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] |
| Pancreatic | Tumor suppressor | mRNA expression down-regulated in tumors[313] | |
| ZIP3/SLC39A3 | Pancreatic | Tumor suppressor | Decreased expression in adenocarcinoma[308,313,331,343] |
| Oncogene | Medium to high mRNA expression in multiple human PC cell lines[313] | ||
| ZIP4/SLC39A4 | Hepatocellular | Oncogene | Increased mRNA and protein expression, repressed apoptosis, enhanced cell cycle and migration[308,325,344-346] |
| Pancreatic | Oncogene | Increased expression in PDAC and PC cell lines, link to CREB-miR-373 axis, promotes cancer xenograft growth in nude mice[313,325-327] | |
| Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] | |
| ZIP5/SLC39A5 | Esophageal | Oncogene | Increased expression in ESCC, knockdown in cell lines inhibited migration and invasion[308,334] |
| Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] | |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| ZIP6/LIV-1/SLC39A6 | Pancreatic | Oncogene | Increased expression in tumors and cell lines[313,330] |
| Tumor suppressor | Decreased mRNA expression in tumors[313] | ||
| Hepatocellular | Oncogene | Increased mRNA and protein expression[308,347] | |
| Esophageal | Oncogene | Increased expression in ESCC[308,333] | |
| Colorectal | Oncogene | Increased mRNA expression in tumors[337] | |
| Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] | |
| ZIP7/SLC39A7 | Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| Colorectal | Oncogene | Increased mRNA expression in tumors and CRC cell lines, knockdown inhibits cell growth and induces apoptosis in cell lines[339] | |
| Gastric | Undetermined | Increased mRNA expression, but better patient prognosis[338] | |
| ZIP8/SLC39A8 | Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| ZIP9/SLC39A9 | Colorectal | Oncogene | Increased mRNA expression in tumors[337] |
| Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] | |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| ZIP10/SLC39A10 | Colorectal | Oncogene | Increased mRNA expression in tumors[337] |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| ZIP11/SLC39A11 | Colorectal | Oncogene | Increased mRNA expression in tumors[337] |
| Gastric | Undetermined | Increased mRNA expression in tumors, better patient prognosis[338] | |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| ZIP12/SLC39A12 | Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| ZIP13/SLC39A13 | Gastric | Oncogene | Increased mRNA expression in tumors, worse patient prognosis[338] |
| Pancreatic | Tumor suppressor | Decreased mRNA expression in tumors[313] | |
| Oncogene | Medium to high mRNA expression in multiple human cell lines[313] | ||
| ZIP14/SLC39A14 | Hepatocellular | Tumor suppressor | Decreased expression in hepatoma tissues[308,348] |
| Gastric | Undetermined | Increased mRNA expression in tumors, but better patient prognosis[338] | |
| Tumor suppressor | Decreased mRNA expression in tumors[313] | ||
| Pancreatic | Oncogene | Medium to high mRNA expression in multiple human cell lines[313] |
The mechanisms underlying the effects of dysregulation of zinc homeostasis and zinc transporter expression in cancer are unclear, and are almost certainly tissue and cancer specific as seen in the evidence that zinc transporters appear to clearly act as tumor suppressors in the prostate gland but as oncogenes in breast cancer. Less is known in GI tract cancers, and that data is controversial (see conflicting PC and EC studies above) but some potential mechanisms of action are listed below.
Zinc signaling within cells: Changes in intracellular concentration can lead to it acting as a second messenger for external signals, including activation of mitogen-activated protein kinase (MAPK), extra-cellular signal-related kinase (ERK) and the c-Jun N-terminal kinase (JNK) pathways that can act to phosphorylate proteins involved in regulating cell proliferation, differentiation and apoptosis[306,322]. This has been best studied in breast cancer where ZIP-mediated signaling pathways promote cell proliferation and metastasis in luminal and tamoxifen-resistant breast cancer cells[322]. Overall, zinc is recognized as an important signaling molecule in normal cell functions as well as pathologies such as cancer. How zinc signaling is conveyed from zinc transporters to downstream signaling pathways is still largely unclear.
Regulation of the intestinal stem cell compartment: In normal tissue several zinc transporters are involved in maintenance of Lgr5+ stem cells and Paneth cells, which themselves are important for stem cell maintenance[312]. Further evidence for this mechanism is the role of ZIP7 in resolving ER stress in the intestinal stem cell compartment[340]. Mice lacking ZIP7 in intestinal epithelium triggered ER stress that led to loss of intestinal stem cells and proliferative progenitor cells.
Requirement for zinc in rapidly proliferating cells: Zinc is required for protein structure, catalytic activity, such as for zinc finger transcription factors.
Action of increased cellular zinc on other ion channels: TRP Ca2+ and K+ channels are sensitive to zinc activation[308].
Zinc deficiency: Zinc deficiency leading to disruption of intestinal epithelial barrier function and enhanced colonic inflammation as zinc supplementation has been shown to improve inflammatory bowel disease-related phenotypes in animal models.
In contrast with other ion channels discussed in this review drugs targeting various zinc transporters have been slow in development. Myers[341] and Bin et al[342] discuss some of the challenges to drug development represented by zinc transporter structure. One of these, ZnT8, which is especially associated with type-2 diabetes is the subject of current drug development research. Potentially, knowledge gained from targeting ZnT8 could lead to targeting of other transporters such as ZIP4 in PC. One therapeutic area that is immediately available is zinc supplementation, also discussed by Myers and Bin et al[342]. For conditions, including cancers, that demonstrate zinc deficiency, zinc supplementation may have immediate therapeutic value. This has been demonstrated for inflammatory bowel disease and may have utility for zinc-deficient cancers. See Table 5 for a full listing of zinc transporters and their role in GI cancers.
Ion channels play an essential role in the GI tract, mediating a range of cellular and tissue processes. They are also commonly dysregulated in major GI malignancies such as CRC, HCC, PC, GC and EC. In these cancers, ion channels modulate many of the well-known hallmarks of cancer, with increasing evidence that ion channels are important for the regulation of tissue and cancer stem cells. The influence of ion channels on cancer cell processes has led to cancer being described as a channelopathy. For a summary of mechanisms of selected ion channels in GI cancer see Figure 3. Notably, ion channels represent potentially important clinical targets for several reasons.
First, ion channels and transporters are predominantly found in the PM of the lumens of GI tract organs thus the majority of ion channels should be accessible to therapeutic drugs.
Second, the structures of all of the major families of ion channels found in the GI tract are known and their functions have been well-characterized, primarily due to studies prompted by dysregulation of these ion channels outside the GI tract, e.g., CFTR in CF lung pathology, thus they should be readily amenable to new drug design and preclinical and clinical testing.
Third, drugs are currently used to target several ion channels for disorders outside the GI tract e.g., retigabine for KCNQ-deficiency and many other examples, thus these drugs can be repurposed for clinical use in the GI tract. Current examples of drug repurposing in the GI tract include CFTR potentiators, activators, correctors, and amplifiers such as for patients with specific CFTR mutations. These include Ivacaftor (potentiator for > 25 CF-causing mutations, including G551D), and Tezacaftor and Lumacaftor (correctors for patients with F508delta mutations which account for aaproximately 70% of CF patients). These drugs have been shown to be effective in ameliorating lung pathology in CF patients and are now being used to treat several GI pathologies in CF patients. For example, Ivacaftor is being used to treat pancreatitis, and intestinal inflammation (including producing an improvement in gut microbiota) and Lumacaftor has been shown to improve intestinal bicarbonate permeability. As CF patients are highly susceptible to the development of CRC, these drugs may be readily repurposed for prevention and treatment of CRC, likely in combination with other standard therapy. Data generated by the repurposing efforts underway for non-cancer therapy will provide support for the next step into the cancer clinic.
Fourth, further research into the mechanisms of action of various ion channels, including the rapidly growing utility of bioinformatics analysis, can lead to greater drug repurposing strategies. For example, through a bioinformatics approach, tricyclic antidepressants were recently and rapidly repurposed by the FDA for use in treatment of small cell lung carcinoma[349]. In other cases, research into mechanisms of drug action reveals ion channels as novel mediators. For example, it has long been known that daily aspirin is protective against CRC, but only recently has it been determined that the mechanism of this effect is likely through the remodeling of the SOCE Ca2+ channel[256]. Further understanding of the canonical and non-canonical roles that ion channels play in cancer development can lead to further repurposing of drugs to treat specific GI cancers[350]. This is especially true as precision cancer medicine embraces combinatorial treatment modalities.
Fifth, ion channels and transporters can provide novel cancer biomarkers with diagnostic and prognostic implications, e.g., KCNQ1 in later stage CRC, where patients who maintain high expression of KCNQ1 show better disease free survival at stage II and III CRC[38], and a 23-month survival advantage for stage IV CRC patients following hepatic resection[37]; CFTR in CF-related CRC[122-126]; hERG1 in several GI cancers[59-74]. and zinc transporters such as ZIP4 in PC[325-329]. For example in CRC, hERG1 is not expressed in normal colon mucosa but is upregulated in adenocarcinoma, with its highest expression in CRC metastasis. It is noted that stereoselective hERG1 channel blockers for treatment of cardiac arrhythmias have been used in the clinic for several decades.
Sixth, there is research data linking several ion channels to regulation of the stem cell compartment in GI tract organs. This is especially true for the intestinal tract. For example, both CFTR[93,112] and KCNQ1 (R.Cormier and P.Scott, unpublished studies) are expressed in the stem cell compartment of the colon, where they have been shown to influence stem call capacity in mouse organoids models[37,128], as well as regulating the expression of stem cell related genes, again in transgenic mouse models[37,128]. As the intestinal stem cell is thought to be the precursor for the intestinal cancer stem cell, better understanding of the underlying mechanisms of how ion channels such as CFTR and KCNQ1 may regulate stem cell function will be very important. A key tool in this research and a technical advance in therapeutic development has been the creation of patient-derived tissue and cancer organoid surrogate models[136], e.g., that have been used to test the function of CFTR[137,138], as well as the efficacy of CFTR modulator drugs in CF patient-derived rectal organoids[208]. The same strategy of biobanking of patient-derived organoids is currently used to test patient-specific CRC treatment protocols that can include ion channel modulator drugs.
Seventh, as patient genomic sequencing efforts increase, more has become known about specific germline genomic variants in ion channel genes and potential susceptibility to GI tract cancers. For example, CF patients who are homozygous for CFTR mutations are at a significantly heightened risk for developing early onset CRC. What is the risk for CRC for heterozygous carriers of CFTR mutations, a group that represents more than 10 million individuals in the United States alone? CRC patients whose cancers show reduced expression of CFTR demonstrate worse CRC disease prognosis[128]. Current ongoing studies of CRC risk in CFTR carriers may help inform on the lifetime risk of CRC in these patients, potentially leading to earlier screening and chemoprevention.
A final comment here, as discussed by Humphries et al[351], is that while there exists great promise in targeting ion channels in human diseases there remains significant challenges, especially related to target specificity and off-target toxicity, before development of ion channel targeting can lead to more widespread effective personalized cancer treatment.
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
Manuscript source: Unsolicited manuscript
P-Reviewer: Camacho J, Ding MX, Sun XT S-Editor: Yan JP L-Editor: A E-Editor: Zhang YL
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13300] [Cited by in F6Publishing: 14452] [Article Influence: 2890.4] [Reference Citation Analysis (2)] |
| 2. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19834] [Cited by in F6Publishing: 18704] [Article Influence: 779.3] [Reference Citation Analysis (0)] |
| 3. | Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42993] [Article Influence: 3307.2] [Reference Citation Analysis (4)] |
| 4. | Litan A, Langhans SA. Cancer as a channelopathy: Ion channels and pumps in tumor development and progression. Front Cell Neurosci. 2015;9:86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 198] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 5. | Prevarskaya N, Skryma R, Shuba Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol Rev. 2018;98:559-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 263] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 6. | Berridge MJ. Calcium microdomains: Organization and function. Cell Calcium. 2006;40:405-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 382] [Cited by in F6Publishing: 376] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 7. | McFerrin MB, Sontheimer H. A role for ion channels in glioma cell invasion. Neuron Glia Biol. 2006;2:39-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 8. | Ruan YC, Wang Y, Da Silva N, Kim B, Diao RY, Hill E, Brown D, Chan HC, Breton S. CFTR interacts with ZO-1 to regulate tight junction assembly and epithelial differentiation through the ZONAB pathway. J Cell Sci. 2014;127:4396-4408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 9. | Bates E. Ion channels in development and cancer. Annu Rev Cell Dev Biol. 2015;31:231-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 137] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 10. | Fiorio Pla A, Munaron L. Functional properties of ion channels and transporters in tumour vascularization. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159-173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 326] [Cited by in F6Publishing: 343] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 12. | Djamgoz MB, Coombes RC, Schwab A. Ion transport and cancer: From initiation to metastasis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130092. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 13. | Pedersen SF, Stock C. Ion channels and transporters in cancer: Pathophysiology, regulation, and clinical potential. Cancer Res. 2013;73:1658-1661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 14. | Stock C, Schwab A. Ion channels and transporters in metastasis. Biochim Biophys Acta. 2015;1848:2638-2646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Schwab A, Stock C. Ion channels and transporters in tumour cell migration and invasion. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 16. | Pardo LA, Contreras-Jurado C, Zientkowska M, Alves F, Stühmer W. Role of voltage-gated potassium channels in cancer. J Membr Biol. 2005;205:115-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 150] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 17. | Comes N, Serrano-Albarrás A, Capera J, Serrano-Novillo C, Condom E, Ramón Y Cajal S, Ferreres JC, Felipe A. Involvement of potassium channels in the progression of cancer to a more malignant phenotype. Biochim Biophys Acta. 2015;1848:2477-2492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 18. | Pardo LA, Stühmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 335] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 19. | Kondratskyi A, Kondratska K, Skryma R, Prevarskaya N. Ion channels in the regulation of apoptosis. Biochim Biophys Acta. 2015;1848:2532-2546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 20. | Abdul Kadir L, Stacey M, Barrett-Jolley R. Emerging Roles of the Membrane Potential: Action Beyond the Action Potential. Front Physiol. 2018;9:1661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 21. | Liin SI, Barro-Soria R, Larsson HP. The KCNQ1 channel - remarkable flexibility in gating allows for functional versatility. J Physiol. 2015;593:2605-2615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, Jentsch TJ. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403:196-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 362] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 23. | Nanda Kumar NS, Singh SK, Rajendran VM. Mucosal potassium efflux mediated via Kcnn4 channels provides the driving force for electrogenic anion secretion in colon. Am J Physiol Gastrointest Liver Physiol. 2010;299:G707-G714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Vallon V, Grahammer F, Volkl H, Sandu CD, Richter K, Rexhepaj R, Gerlach U, Rong Q, Pfeifer K, Lang F. KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proc Natl Acad Sci U S A. 2005;102:17864-17869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 148] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Julio-Kalajzić F, Villanueva S, Burgos J, Ojeda M, Cid LP, Jentsch TJ, Sepúlveda FV. K2P TASK-2 and KCNQ1-KCNE3 K+ channels are major players contributing to intestinal anion and fluid secretion. J Physiol. 2018;596:393-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 26. | Winbo A, Sandström O, Palmqvist R, Rydberg A. Iron-deficiency anaemia, gastric hyperplasia, and elevated gastrin levels due to potassium channel dysfunction in the Jervell and Lange-Nielsen Syndrome. Cardiol Young. 2013;23:325-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: A mutation update. Hum Mutat. 2009;30:1486-1511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 318] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 28. | Asad-Ur-Rahman FN, Hughes L, Talha Khan M, Khalid Hasan M, Inayat I. Long QT Syndrome and Duodenal Ampullary Adenoma: A New Association. ACG Case Rep J. 2016;3:e163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 29. | Rice KS, Dickson G, Lane M, Crawford J, Chung SK, Rees MI, Shelling AN, Love DR, Skinner JR. Elevated serum gastrin levels in Jervell and Lange-Nielsen syndrome: A marker of severe KCNQ1 dysfunction? Heart Rhythm. 2011;8:551-554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Nakano Y, Shimizu W. Genetics of long-QT syndrome. J Hum Genet. 2016;61:51-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 31. | Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD, Brandenburg SA, Litzi TJ, Bunton TE, Limb C, Francis H, Gorelikow M, Gu H, Washington K, Argani P, Goldenring JR, Coffey RJ, Feinberg AP. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest. 2000;106:1447-1455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 215] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Elso CM, Lu X, Culiat CT, Rutledge JC, Cacheiro NL, Generoso WM, Stubbs LJ. Heightened susceptibility to chronic gastritis, hyperplasia and metaplasia in Kcnq1 mutant mice. Hum Mol Genet. 2004;13:2813-2821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, Collier LS, Powers S, Oberg AL, Asmann YW, Thibodeau SN, Tessarollo L, Copeland NG, Jenkins NA, Cormier RT, Largaespada DA. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747-1750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 34. | March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M, van der Weyden L, Berns A, Gadiot J, Uren A, Kemp R, Arends MJ, Wessels LF, Winton DJ, Adams DJ. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202-1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 35. | Takeda H, Wei Z, Koso H, Rust AG, Yew CC, Mann MB, Ward JM, Adams DJ, Copeland NG, Jenkins NA. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet. 2015;47:142-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 36. | Morris SM, Davison J, Carter KT, O'Leary RM, Trobridge P, Knoblaugh SE, Myeroff LL, Markowitz SD, Brett BT, Scheetz TE, Dupuy AJ, Starr TK, Grady WM. Transposon mutagenesis identifies candidate genes that cooperate with loss of transforming growth factor-beta signaling in mouse intestinal neoplasms. Int J Cancer. 2017;140:853-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Than BL, Goos JA, Sarver AL, O'Sullivan MG, Rod A, Starr TK, Fijneman RJ, Meijer GA, Zhao L, Zhang Y, Largaespada DA, Scott PM, Cormier RT. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene. 2014;33:3861-3868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 38. | den Uil SH, Coupé VM, Linnekamp JF, van den Broek E, Goos JA, Delis-van Diemen PM, Belt EJ, van Grieken NC, Scott PM, Vermeulen L, Medema JP, Bril H, Stockmann HB, Cormier RT, Meijer GA, Fijneman RJ. Loss of KCNQ1 expression in stage II and stage III colon cancer is a strong prognostic factor for disease recurrence. Br J Cancer. 2016;115:1565-1574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Pérez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grützmann R, Aust D, Rümmele P, Knösel T, Herd C, Stemple DL, Kettleborough R, Brosnan JA, Li A, Morgan R, Knight S, Yu J, Stegeman S, Collier LS, ten Hoeve JJ, de Ridder J, Klein AP, Goggins M, Hruban RH, Chang DK, Biankin AV, Grimmond SM; Australian Pancreatic Cancer Genome Initiative, Wessels LF, Wood SA, Iacobuzio-Donahue CA, Pilarsky C, Largaespada DA, Adams DJ, Tuveson DA. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 255] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 40. | Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ; Australian Pancreatic Cancer Genome Initiative, Chang DK, Biankin AV, Waddell N, Kassahn KS, Grimmond SM, Rust AG, Adams DJ, Jenkins NA, Copeland NG. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:5934-5941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 176] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 41. | Bard-Chapeau EA, Nguyen AT, Rust AG, Sayadi A, Lee P, Chua BQ, New LS, de Jong J, Ward JM, Chin CK, Chew V, Toh HC, Abastado JP, Benoukraf T, Soong R, Bard FA, Dupuy AJ, Johnson RL, Radda GK, Chan EC, Wessels LF, Adams DJ, Jenkins NA, Copeland NG. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46:24-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 42. | Takeda H, Rust AG, Ward JM, Yew CC, Jenkins NA, Copeland NG. Sleeping Beauty transposon mutagenesis identifies genes that cooperate with mutant Smad4 in gastric cancer development. Proc Natl Acad Sci U S A. 2016;113:E2057-E2065. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 43. | Zhang J, Zhao Z, Zu C, Hu H, Shen H, Zhang M, Wang J. Atrial natriuretic peptide modulates the proliferation of human gastric cancer cells via KCNQ1 expression. Oncol Lett. 2013;6:407-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 44. | Fan H, Zhang M, Liu W. Hypermethylated KCNQ1 acts as a tumor suppressor in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;503:3100-3107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Rapetti-Mauss R, Bustos V, Thomas W, McBryan J, Harvey H, Lajczak N, Madden SF, Pellissier B, Borgese F, Soriani O, Harvey BJ. Bidirectional KCNQ1:β-catenin interaction drives colorectal cancer cell differentiation. Proc Natl Acad Sci U S A. 2017;114:4159-4164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 46. | Preston P, Wartosch L, Günzel D, Fromm M, Kongsuphol P, Ousingsawat J, Kunzelmann K, Barhanin J, Warth R, Jentsch TJ. Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl- transport. J Biol Chem. 2010;285:7165-7175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 47. | Wilmes J, Haddad-Tóvolli R, Alesutan I, Munoz C, Sopjani M, Pelzl L, Bogatikov E, Fedele G, Faggio C, Seebohm G, Föller M, Lang F. Regulation of KCNQ1/KCNE1 by β-catenin. Mol Membr Biol. 2012;29:87-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Kanduri C. Kcnq1ot1: A chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 49. | Bian Y, Gao G, Zhang Q, Qian H, Yu L, Yao N, Qian J, Liu B, Qian X. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2019;20:886-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 50. | Yang F, Wen S, Zhang Y, Xu Y, Lv H, Zhu Y, Wang M, Su P, Huang C, Tian Z. Identifying potential metastasis-related long non-coding RNAs, microRNAs, and message RNAs in the esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:13202-13215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 51. | Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol. 2018;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 52. | Sunamura N, Ohira T, Kataoka M, Inaoka D, Tanabe H, Nakayama Y, Oshimura M, Kugoh H. Regulation of functional KCNQ1OT1 lncRNA by β-catenin. Sci Rep. 2016;6:20690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 53. | Gunthorpe MJ, Large CH, Sankar R. The mechanism of action of retigabine (ezogabine), a first-in-class K+ channel opener for the treatment of epilepsy. Epilepsia. 2012;53:412-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 54. | Roepke TK, Purtell K, King EC, La Perle KM, Lerner DJ, Abbott GW. Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS One. 2010;5:e11451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 55. | Yanglin P, Lina Z, Zhiguo L, Na L, Haifeng J, Guoyun Z, Jie L, Jun W, Tao L, Li S, Taidong Q, Jianhong W, Daiming F. KCNE2, a down-regulated gene identified by in silico analysis, suppressed proliferation of gastric cancer cells. Cancer Lett. 2007;246:129-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Kuwahara N, Kitazawa R, Fujiishi K, Nagai Y, Haraguchi R, Kitazawa S. Gastric adenocarcinoma arising in gastritis cystica profunda presenting with selective loss of KCNE2 expression. World J Gastroenterol. 2013;19:1314-1317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 15] [Cited by in F6Publishing: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Li X, Cai H, Zheng W, Tong M, Li H, Ao L, Li J, Hong G, Li M, Guan Q, Yang S, Yang D, Lin X, Guo Z. An individualized prognostic signature for gastric cancer patients treated with 5-Fluorouracil-based chemotherapy and distinct multi-omics characteristics of prognostic groups. Oncotarget. 2016;7:8743-8755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 58. | Abbott GW, Roepke TK. KCNE2 and gastric cancer: Bench to bedside. Oncotarget. 2016;7:17286-17287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 59. | Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H, Bencini L, Calistri M, Messerini L, Scatizzi M, Moretti R, Wanke E, Olivotto M, Mugnai G, Arcangeli A. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64:606-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 60. | Crociani O, Zanieri F, Pillozzi S, Lastraioli E, Stefanini M, Fiore A, Fortunato A, D'Amico M, Masselli M, De Lorenzo E, Gasparoli L, Chiu M, Bussolati O, Becchetti A, Arcangeli A. hERG1 channels modulate integrin signaling to trigger angiogenesis and tumor progression in colorectal cancer. Sci Rep. 2013;3:3308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 61. | Fiore A, Carraresi L, Morabito A, Polvani S, Fortunato A, Lastraioli E, Femia AP, De Lorenzo E, Caderni G, Arcangeli A. Characterization of hERG1 channel role in mouse colorectal carcinogenesis. Cancer Med. 2013;2:583-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Lang F, Stournaras C. Ion channels in cancer: Future perspectives and clinical potential. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 63. | Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang W, Wang B, Yang L, Xu H, Zhang G, Xu Z. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget. 2014;5:5832-5844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | Manoli S, Coppola S, Duranti C, Lulli M, Magni L, Kuppalu N, Nielsen N, Schmidt T, Schwab A, Becchetti A, Arcangeli A. The Activity of Kv 11.1 Potassium Channel Modulates F-Actin Organization During Cell Migration of Pancreatic Ductal Adenocarcinoma Cells. Cancers (Basel). 2019;11:pii: E135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 65. | Arcangeli A, Crociani O, Bencini L. Interaction of tumour cells with their microenvironment: Ion channels and cell adhesion molecules. A focus on pancreatic cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Lastraioli E, Perrone G, Sette A, Fiore A, Crociani O, Manoli S, D'Amico M, Masselli M, Iorio J, Callea M, Borzomati D, Nappo G, Bartolozzi F, Santini D, Bencini L, Farsi M, Boni L, Di Costanzo F, Schwab A, Onetti Muda A, Coppola R, Arcangeli A. hERG1 channels drive tumour malignancy and may serve as prognostic factor in pancreatic ductal adenocarcinoma. Br J Cancer. 2015;112:1076-1087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 67. | Ding XW, Luo HS, Luo B, Xu DQ, Gao S. Overexpression of hERG1 in resected esophageal squamous cell carcinomas: A marker for poor prognosis. J Surg Oncol. 2008;97:57-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 68. | Xia J, Wang H, Li S, Wu Q, Sun L, Huang H, Zeng M. Ion channels or aquaporins as novel molecular targets in gastric cancer. Mol Cancer. 2017;16:54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 69. | Lastraioli E, Taddei A, Messerini L, Comin CE, Festini M, Giannelli M, Tomezzoli A, Paglierani M, Mugnai G, De Manzoni G, Bechi P, Arcangeli A. hERG1 channels in human esophagus: Evidence for their aberrant expression in the malignant progression of Barrett's esophagus. J Cell Physiol. 2006;209:398-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 70. | Lastraioli E, Lottini T, Iorio J, Freschi G, Fazi M, Duranti C, Carraresi L, Messerini L, Taddei A, Ringressi MN, Salemme M, Villanacci V, Vindigni C, Tomezzoli A, La Mendola R, Bencivenga M, Compagnoni B, Chiudinelli M, Saragoni L, Manzi I, De Manzoni G, Bechi P, Boni L, Arcangeli A. hERG1 behaves as biomarker of progression to adenocarcinoma in Barrett's esophagus and can be exploited for a novel endoscopic surveillance. Oncotarget. 2016;7:59535-59547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 71. | Ding XW, Yang WB, Gao S, Wang W, Li Z, Hu WM, Li JJ, Luo HS. Prognostic significance of hERG1 expression in gastric cancer. Dig Dis Sci. 2010;55:1004-1010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 72. | Crociani O, Lastraioli E, Boni L, Pillozzi S, Romoli MR, D'Amico M, Stefanini M, Crescioli S, Masi A, Taddei A, Bencini L, Bernini M, Farsi M, Beghelli S, Scarpa A, Messerini L, Tomezzoli A, Vindigni C, Morgagni P, Saragoni L, Giommoni E, Gasperoni S, Di Costanzo F, Roviello F, De Manzoni G, Bechi P, Arcangeli A. hERG1 channels regulate VEGF-A secretion in human gastric cancer: Clinicopathological correlations and therapeutical implications. Clin Cancer Res. 2014;20:1502-1512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 73. | Arcangeli A, Crociani O, Lastraioli E, Masi A, Pillozzi S, Becchetti A. Targeting ion channels in cancer: A novel frontier in antineoplastic therapy. Curr Med Chem. 2009;16:66-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 74. | Arcangeli A, Becchetti A. hERG Channels: From Antitargets to Novel Targets for Cancer Therapy. Clin Cancer Res. 2017;23:3-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Hemmerlein B, Weseloh RM, Mello de Queiroz F, Knötgen H, Sánchez A, Rubio ME, Martin S, Schliephacke T, Jenke M, Heinz-Joachim-Radzun, Stühmer W, Pardo LA. Overexpression of Eag1 potassium channels in clinical tumours. Mol Cancer. 2006;5:41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 200] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 76. | Ousingsawat J, Spitzner M, Puntheeranurak S, Terracciano L, Tornillo L, Bubendorf L, Kunzelmann K, Schreiber R. Expression of voltage-gated potassium channels in human and mouse colonic carcinoma. Clin Cancer Res. 2007;13:824-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 105] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 77. | Chávez-López MG, Zúñiga-García V, Pérez-Carreón JI, Avalos-Fuentes A, Escobar Y, Camacho J. Eag1 channels as potential early-stage biomarkers of hepatocellular carcinoma. Biologics. 2016;10:139-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 78. | Han Y, Shi Y, Han Z, Sun L, Fan D. Detection of potassium currents and regulation of multidrug resistance by potassium channels in human gastric cancer cells. Cell Biol Int. 2007;31:741-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 79. | Bielanska J, Hernández-Losa J, Pérez-Verdaguer M, Moline T, Somoza R, Ramón Y Cajal S, Condom E, Ferreres JC, Felipe A. Voltage-dependent potassium channels Kv1.3 and Kv1.5 in human cancer. Curr Cancer Drug Targets. 2009;9:904-914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 80. | Lan M, Shi Y, Han Z, Hao Z, Pan Y, Liu N, Guo C, Hong L, Wang J, Qiao T, Fan D. Expression of delayed rectifier potassium channels and their possible roles in proliferation of human gastric cancer cells. Cancer Biol Ther. 2005;4:1342-1347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 81. | Kim HJ, Jang SH, Jeong YA, Ryu PD, Kim DY, Lee SY. Involvement of Kv4.1 K(+) channels in gastric cancer cell proliferation. Biol Pharm Bull. 2010;33:1754-1757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 82. | Brevet M, Fucks D, Chatelain D, Regimbeau JM, Delcenserie R, Sevestre H, Ouadid-Ahidouch H. Deregulation of 2 potassium channels in pancreas adenocarcinomas: Implication of KV1.3 gene promoter methylation. Pancreas. 2009;38:649-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 83. | Lai W, Liu L, Zeng Y, Wu H, Xu H, Chen S, Chu Z. KCNN4 channels participate in the EMT induced by PRL-3 in colorectal cancer. Med Oncol. 2013;30:566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Jäger H, Dreker T, Buck A, Giehl K, Gress T, Grissmer S. Blockage of intermediate-conductance Ca2+-activated K+ channels inhibit human pancreatic cancer cell growth in vitro. Mol Pharmacol. 2004;65:630-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 151] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 85. | Lee JH, Park JW, Byun JK, Kim HK, Ryu PD, Lee SY, Kim DY. Silencing of voltage-gated potassium channel KV9.3 inhibits proliferation in human colon and lung carcinoma cells. Oncotarget. 2015;6:8132-8143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 86. | Pei L, Wiser O, Slavin A, Mu D, Powers S, Jan LY, Hoey T. Oncogenic potential of TASK3 (Kcnk9) depends on K+ channel function. Proc Natl Acad Sci USA. 2003;100:7803-7807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 128] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 87. | Mu D, Chen L, Zhang X, See LH, Koch CM, Yen C, Tong JJ, Spiegel L, Nguyen KC, Servoss A, Peng Y, Pei L, Marks JR, Lowe S, Hoey T, Jan LY, McCombie WR, Wigler MH, Powers S. Genomic amplification and oncogenic properties of the KCNK9 potassium channel gene. Cancer Cell. 2003;3:297-302. [PubMed] [Cited in This Article: ] |
| 88. | He T, Wang C, Zhang M, Zhang X, Zheng S, Linghu E, Guo M. Epigenetic regulation of voltage-gated potassium ion channel molecule Kv1.3 in mechanisms of colorectal cancer. Discov Med. 2017;23:155-162. [PubMed] [Cited in This Article: ] |
| 89. | Ouadid-Ahidouch H, Rodat-Despoix L, Matifat F, Morin G, Ahidouch A. DNA methylation of channel-related genes in cancers. Biochim Biophys Acta. 2015;1848:2621-2628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 90. | Xu E, Gu J, Hawk ET, Wang KK, Lai M, Huang M, Ajani J, Wu X. Genome-wide methylation analysis shows similar patterns in Barrett's esophagus and esophageal adenocarcinoma. Carcinogenesis. 2013;34:2750-2756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 91. | Lastraioli E, Iorio J, Arcangeli A. Ion channel expression as promising cancer biomarker. Biochim Biophys Acta. 2015;1848:2685-2702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 92. | Crites KS, Morin G, Orlando V, Patey N, Cantin C, Martel J, Brochiero E, Mailhot G. CFTR Knockdown induces proinflammatory changes in intestinal epithelial cells. J Inflamm (Lond). 2015;12:62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 93. | Strubberg AM, Liu J, Walker NM, Stefanski CD, MacLeod RJ, Magness ST, Clarke LL. Cftr Modulates Wnt/β-Catenin Signaling and Stem Cell Proliferation in Murine Intestine. Cell Mol Gastroenterol Hepatol. 2017;5:253-271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 94. | Vernocchi P, Del Chierico F, Russo A, Majo F, Rossitto M, Valerio M, Casadei L, La Storia A, De Filippis F, Rizzo C, Manetti C, Paci P, Ercolini D, Marini F, Fiscarelli EV, Dallapiccola B, Lucidi V, Miccheli A, Putignani L. Gut microbiota signatures in cystic fibrosis: Loss of host CFTR function drives the microbiota enterophenotype. PLoS One. 2018;13:e0208171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 95. | Burke DG, Fouhy F, Harrison MJ, Rea MC, Cotter PD, O'Sullivan O, Stanton C, Hill C, Shanahan F, Plant BJ, Ross RP. The altered gut microbiota in adults with cystic fibrosis. BMC Microbiol. 2017;17:58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 96. | Kleme ML, Sané AT, Garofalo C, Levy E. Targeted CFTR gene disruption with zinc-finger nucleases in human intestinal epithelial cells induces oxidative stress and inflammation. Int J Biochem Cell Biol. 2016;74:84-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 97. | Kleme ML, Sané A, Garofalo C, Seidman E, Brochiero E, Berthiaume Y, Levy E. CFTR Deletion Confers Mitochondrial Dysfunction and Disrupts Lipid Homeostasis in Intestinal Epithelial Cells. Nutrients. 2018;10:pii: E836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 98. | Xia X, Wang J, Liu Y, Yue M. Lower Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Promotes the Proliferation and Migration of Endometrial Carcinoma. Med Sci Monit. 2017;23:966-974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 99. | Liou TG. The Clinical Biology of Cystic Fibrosis Transmembrane Regulator Protein: Its Role and Function in Extrapulmonary Disease. Chest. 2019;155:605-616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 100. | Zhang JT, Wang Y, Chen JJ, Zhang XH, Dong JD, Tsang LL, Huang XR, Cai Z, Lan HY, Jiang XH, Chan HC. Defective CFTR leads to aberrant β-catenin activation and kidney fibrosis. Sci Rep. 2017;7:5233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 101. | Zhang J, Wang Y, Jiang X, Chan HC. Cystic fibrosis transmembrane conductance regulator-emerging regulator of cancer. Cell Mol Life Sci. 2018;75:1737-1756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 102. | Villella VR, Venerando A, Cozza G, Esposito S, Ferrari E, Monzani R, Spinella MC, Oikonomou V, Renga G, Tosco A, Rossin F, Guido S, Silano M, Garaci E, Chao YK, Grimm C, Luciani A, Romani L, Piacentini M, Raia V, Kroemer G, Maiuri L. A pathogenic role for cystic fibrosis transmembrane conductance regulator in celiac disease. EMBO J. 2019;38:pii: e100101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 103. | Garg M, Ooi CY. The Enigmatic Gut in Cystic Fibrosis: Linking Inflammation, Dysbiosis, and the Increased Risk of Malignancy. Curr Gastroenterol Rep. 2017;19:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 104. | Gelfond D, Heltshe S, Ma C, Rowe SM, Frederick C, Uluer A, Sicilian L, Konstan M, Tullis E, Roach RN, Griffin K, Joseloff E, Borowitz D. Impact of CFTR Modulation on Intestinal pH, Motility, and Clinical Outcomes in Patients With Cystic Fibrosis and the G551D Mutation. Clin Transl Gastroenterol. 2017;8:e81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 105. | Romi H, Cohen I, Landau D, Alkrinawi S, Yerushalmi B, Hershkovitz R, Newman-Heiman N, Cutting GR, Ofir R, Sivan S, Birk OS. Meconium ileus caused by mutations in GUCY2C, encoding the CFTR-activating guanylate cyclase 2C. Am J Hum Genet. 2012;90:893-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 106. | Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066-1073. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5288] [Cited by in F6Publishing: 5081] [Article Influence: 145.2] [Reference Citation Analysis (0)] |
| 107. | Maitra A. Cystic Fibrosis. In: Kumar V, Abbas AK, Aster JC, eds. Robbins and Cotran Pathologic Basis of Disease. 9th ed. Elsevier, 2015. [Cited in This Article: ] |
| 108. | Sorscher EJ. Cystic Fibrosis. In: Kasper D, Fauci A, Hauser S, Longo D, Jameson JL, Loscalzo J, eds. Harrison's Principles of Internal Medicine. 19th ed. New York, NY: McGraw-Hill Education, 2015. [Cited in This Article: ] |
| 109. | Tse CM, Yin J, Singh V, Sarker R, Lin R, Verkman AS, Turner JR, Donowitz M. cAMP Stimulates SLC26A3 Activity in Human Colon by a CFTR-Dependent Mechanism That Does Not Require CFTR Activity. Cell Mol Gastroenterol Hepatol. 2019;7:641-653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 110. | Li C, Naren AP. Macromolecular complexes of cystic fibrosis transmembrane conductance regulator and its interacting partners. Pharmacol Ther. 2005;108:208-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 111. | Guggino WB, Stanton BA. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat Rev Mol Cell Biol. 2006;7:426-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 332] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 112. | Jakab RL, Collaco AM, Ameen NA. Physiological relevance of cell-specific distribution patterns of CFTR, NKCC1, NBCe1, and NHE3 along the crypt-villus axis in the intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G82-G98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 113. | Jakab RL, Collaco AM, Ameen NA. Characterization of CFTR High Expresser cells in the intestine. Am J Physiol Gastrointest Liver Physiol. 2013;305:G453-G465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 114. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3854] [Cited by in F6Publishing: 4012] [Article Influence: 236.0] [Reference Citation Analysis (0)] |
| 115. | Moskowitz SM, Chmiel JF, Sternen DL, Cheng E, Gibson RL, Marshall SG, Cutting GR. Clinical practice and genetic counseling for cystic fibrosis and CFTR-related disorders. Genet Med. 2008;10:851-868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 116. | Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918-951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1180] [Cited by in F6Publishing: 1129] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 117. | Castellani C, Assael BM. Cystic fibrosis: A clinical view. Cell Mol Life Sci. 2017;74:129-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 137] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 118. | Wilschanski M, Novak I. The cystic fibrosis of exocrine pancreas. Cold Spring Harb Perspect Med. 2013;3:a009746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 119. | Hou Y, Guan X, Yang Z, Li C. Emerging role of cystic fibrosis transmembrane conductance regulator - an epithelial chloride channel in gastrointestinal cancers. World J Gastrointest Oncol. 2016;8:282-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 120. | De Lisle RC, Borowitz D. The cystic fibrosis intestine. Cold Spring Harb Perspect Med. 2013;3:a009753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 121. | Cystic Fibrosis Foundation. Patient Registry, 2019. Available from: https://www.cff.org. [Cited in This Article: ] |
| 122. | Maisonneuve P, Marshall BC, Knapp EA, Lowenfels AB. Cancer risk in cystic fibrosis: A 20-year nationwide study from the United States. J Natl Cancer Inst. 2013;105:122-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 204] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 123. | Yamada A, Komaki Y, Komaki F, Micic D, Zullow S, Sakuraba A. Risk of gastrointestinal cancers in patients with cystic fibrosis: A systematic review and meta-analysis. Lancet Oncol. 2018;19:758-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 124. | Niccum DE, Billings JL, Dunitz JM, Khoruts A. Colonoscopic screening shows increased early incidence and progression of adenomas in cystic fibrosis. J Cyst Fibros. 2016;15:548-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 125. | Billings JL, Dunitz JM, McAllister S, Herzog T, Bobr A, Khoruts A. Early colon screening of adult patients with cystic fibrosis reveals high incidence of adenomatous colon polyps. J Clin Gastroenterol. 2014;48:e85-e88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 126. | Hadjiliadis D, Khoruts A, Zauber AG, Hempstead SE, Maisonneuve P, Lowenfels AB; Cystic Fibrosis Colorectal Cancer Screening Task Force. Cystic Fibrosis Colorectal Cancer Screening Consensus Recommendations. Gastroenterology. 2018;154:736-745.e14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 127. | Abbott KL, Nyre ET, Abrahante J, Ho YY, Isaksson Vogel R, Starr TK. The Candidate Cancer Gene Database: A database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43:D844-D848. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 128. | Than BL, Linnekamp JF, Starr TK, Largaespada DA, Rod A, Zhang Y, Bruner V, Abrahante J, Schumann A, Luczak T, Walter J, Niemczyk A, O'Sullivan MG, Medema JP, Fijneman RJ, Meijer GA, Van den Broek E, Hodges CA, Scott PM, Vermeulen L, Cormier RT. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene. 2016;35:4179-4187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 129. | Sun TT, Wang Y, Cheng H, Xiao HZ, Xiang JJ, Zhang JT, Yu SB, Martin TA, Ye L, Tsang LL, Jiang WG, Xiaohua J, Chan HC. Disrupted interaction between CFTR and AF-6/afadin aggravates malignant phenotypes of colon cancer. Biochim Biophys Acta. 2014;1843:618-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 130. | Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK, Lee JU, Lee YM, Kwon SJ, Choi E, Na MJ, Park JY, Kim DS. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011;16:1203-1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 131. | Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J, Wang J, Feng X, Li Y, Yao X, Zhu J. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13:7296-7304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 168] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 132. | Zhao Y, Guo S, Sun J, Huang Z, Zhu T, Zhang H, Gu J, He Y, Wang W, Ma K, Wang J, Yu J. Methylcap-seq reveals novel DNA methylation markers for the diagnosis and recurrence prediction of bladder cancer in a Chinese population. PLoS One. 2012;7:e35175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 133. | van der Heijden AG, Mengual L, Ingelmo-Torres M, Lozano JJ, van Rijt-van de Westerlo CCM, Baixauli M, Geavlete B, Moldoveanud C, Ene C, Dinney CP, Czerniak B, Schalken JA, Kiemeney LALM, Ribal MJ, Witjes JA, Alcaraz A. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics. 2018;10:71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 134. | Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1545] [Cited by in F6Publishing: 1578] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 135. | Li P, Singh J, Sun Y, Ma X, Yuan P. CFTR constrains the differentiation from mouse embryonic stem cells to intestine lineage cells. Biochem Biophys Res Commun. 2019;510:322-328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 136. | Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2253] [Cited by in F6Publishing: 2357] [Article Influence: 181.3] [Reference Citation Analysis (0)] |
| 137. | Liu J, Walker NM, Cook MT, Ootani A, Clarke LL. Functional Cftr in crypt epithelium of organotypic enteroid cultures from murine small intestine. Am J Physiol Cell Physiol. 2012;302:C1492-C1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 138. | Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NW, Bijvelds MJ, Scholte BJ, Nieuwenhuis EE, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19:939-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 659] [Cited by in F6Publishing: 688] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 139. | Berkers G, van Mourik P, Vonk AM, Kruisselbrink E, Dekkers JF, de Winter-de Groot KM, Arets HGM, Marck-van der Wilt REP, Dijkema JS, Vanderschuren MM, Houwen RHJ, Heijerman HGM, van de Graaf EA, Elias SG, Majoor CJ, Koppelman GH, Roukema J, Bakker M, Janssens HM, van der Meer R, Vries RGJ, Clevers HC, de Jonge HR, Beekman JM, van der Ent CK. Rectal Organoids Enable Personalized Treatment of Cystic Fibrosis. Cell Rep. 2019;26:1701-1708.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 177] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 140. | Beekman JM. Individualized medicine using intestinal responses to CFTR potentiators and correctors. Pediatr Pulmonol. 2016;51:S23-S34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 141. | McHugh DR, Steele MS, Valerio DM, Miron A, Mann RJ, LePage DF, Conlon RA, Cotton CU, Drumm ML, Hodges CA. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS One. 2018;13:e0199573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 142. | van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1313] [Cited by in F6Publishing: 1487] [Article Influence: 185.9] [Reference Citation Analysis (0)] |
| 143. | Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, Sato T. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell. 2016;18:827-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 516] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 144. | Dow LE, O'Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161:1539-1552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 324] [Cited by in F6Publishing: 374] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 145. | Liu K, Zhang X, Zhang JT, Tsang LL, Jiang X, Chan HC. Defective CFTR- β-catenin interaction promotes NF-κB nuclear translocation and intestinal inflammation in cystic fibrosis. Oncotarget. 2016;7:64030-64042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 146. | de Sousa E Melo F, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, Rodermond H, Dekker E, van der Loos CM, Pals ST, van de Vijver MJ, Versteeg R, Richel DJ, Vermeulen L, Medema JP. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476-485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 243] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 147. | Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, de Stanchina E, Massagué J. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016;165:45-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 538] [Cited by in F6Publishing: 499] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 148. | Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4659-4665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 848] [Cited by in F6Publishing: 904] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 149. | Gustafsson JK, Ermund A, Ambort D, Johansson ME, Nilsson HE, Thorell K, Hebert H, Sjövall H, Hansson GC. Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J Exp Med. 2012;209:1263-1272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 150. | Sun X, Olivier AK, Yi Y, Pope CE, Hayden HS, Liang B, Sui H, Zhou W, Hager KR, Zhang Y, Liu X, Yan Z, Fisher JT, Keiser NW, Song Y, Tyler SR, Goeken JA, Kinyon JM, Radey MC, Fligg D, Wang X, Xie W, Lynch TJ, Kaminsky PM, Brittnacher MJ, Miller SI, Parekh K, Meyerholz DK, Hoffman LR, Frana T, Stewart ZA, Engelhardt JF. Gastrointestinal pathology in juvenile and adult CFTR-knockout ferrets. Am J Pathol. 2014;184:1309-1322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 151. | Liu J, Walker NM, Ootani A, Strubberg AM, Clarke LL. Defective goblet cell exocytosis contributes to murine cystic fibrosis-associated intestinal disease. J Clin Invest. 2015;125:1056-1068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 152. | De Lisle RC. Disrupted tight junctions in the small intestine of cystic fibrosis mice. Cell Tissue Res. 2014;355:131-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 153. | Broadbent D, Ahmadzai MM, Kammala AK, Yang C, Occhiuto C, Das R, Subramanian H. Roles of NHERF Family of PDZ-Binding Proteins in Regulating GPCR Functions. Adv Immunol. 2017;136:353-385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 154. | Tomas J, Mulet C, Saffarian A, Cavin JB, Ducroc R, Regnault B, Kun Tan C, Duszka K, Burcelin R, Wahli W, Sansonetti PJ, Pédron T. High-fat diet modifies the PPAR-γ pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc Natl Acad Sci U S A. 2016;113:E5934-E5943. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 155. | Munck A. Cystic fibrosis: Evidence for gut inflammation. Int J Biochem Cell Biol. 2014;52:180-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 156. | Lynch SV, Goldfarb KC, Wild YK, Kong W, De Lisle RC, Brodie EL. Cystic fibrosis transmembrane conductance regulator knockout mice exhibit aberrant gastrointestinal microbiota. Gut Microbes. 2013;4:41-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 157. | Norkina O, Burnett TG, De Lisle RC. Bacterial overgrowth in the cystic fibrosis transmembrane conductance regulator null mouse small intestine. Infect Immun. 2004;72:6040-6049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 140] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 158. | Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E, Guarino A. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: A randomised clinical trial. PLoS One. 2014;9:e87796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 156] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 159. | Trigo Salado C, Leo Carnerero E, de la Cruz Ramírez MD. Crohn's disease and cystic fibrosis: There is still a lot to learn. Rev Esp Enferm Dig. 2018;110:835-836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 160. | Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 398] [Cited by in F6Publishing: 456] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 161. | Werlin SL, Benuri-Silbiger I, Kerem E, Adler SN, Goldin E, Zimmerman J, Malka N, Cohen L, Armoni S, Yatzkan-Israelit Y, Bergwerk A, Aviram M, Bentur L, Mussaffi H, Bjarnasson I, Wilschanski M. Evidence of intestinal inflammation in patients with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2010;51:304-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 162. | Norkina O, Kaur S, Ziemer D, De Lisle RC. Inflammation of the cystic fibrosis mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2004;286:G1032-G1041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 115] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 163. | Smyth RL, Croft NM, O'Hea U, Marshall TG, Ferguson A. Intestinal inflammation in cystic fibrosis. Arch Dis Child. 2000;82:394-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 100] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 164. | Dhaliwal J, Leach S, Katz T, Nahidi L, Pang T, Lee JM, Strachan R, Day AS, Jaffe A, Ooi CY. Intestinal inflammation and impact on growth in children with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2015;60:521-526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 165. | Bruzzese E, Raia V, Gaudiello G, Polito G, Buccigrossi V, Formicola V, Guarino A. Intestinal inflammation is a frequent feature of cystic fibrosis and is reduced by probiotic administration. Aliment Pharmacol Ther. 2004;20:813-819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 166. | Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, Gumucio D, Neurath MF, Pasparakis M. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 792] [Cited by in F6Publishing: 900] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 167. | Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, Williams IR, Koval M, Peatman E, Campbell JA, Dermody TS, Nusrat A, Parkos CA. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067-3076. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 374] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 168. | Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, Abraham C, Turner JR. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 317] [Cited by in F6Publishing: 323] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 169. | Bhat AA, Uppada S, Achkar IW, Hashem S, Yadav SK, Shanmugakonar M, Al-Naemi HA, Haris M, Uddin S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front Physiol. 2019;9:1942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 216] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 170. | Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3061] [Cited by in F6Publishing: 3050] [Article Influence: 152.5] [Reference Citation Analysis (0)] |
| 171. | Fiorotto R, Villani A, Kourtidis A, Scirpo R, Amenduni M, Geibel PJ, Cadamuro M, Spirli C, Anastasiadis PZ, Strazzabosco M. The cystic fibrosis transmembrane conductance regulator controls biliary epithelial inflammation and permeability by regulating Src tyrosine kinase activity. Hepatology. 2016;64:2118-2134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 172. | Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, Rupec RA, Gerhard M, Schmid R, Barker N, Clevers H, Lang R, Neumann J, Kirchner T, Taketo MM, van den Brink GR, Sansom OJ, Arkan MC, Greten FR. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 821] [Cited by in F6Publishing: 772] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 173. | Dong J, Jiang X, Zhang X, Liu KS, Zhang J, Chen J, Yu MK, Tsang LL, Chung YW, Wang Y, Zhou WL, Chan HC. Dynamically Regulated CFTR Expression and Its Functional Role in Cutaneous Wound Healing. J Cell Physiol. 2015;230:2049-2058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 174. | Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23:203-214.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 281] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 175. | Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, Chen K, Walker J, McDonald S, Bose R, Ornitz D, Xiong D, You M, Dooling DJ, Watson M, Mardis ER, Wilson RK. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121-1134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 830] [Cited by in F6Publishing: 884] [Article Influence: 73.7] [Reference Citation Analysis (0)] |
| 176. | Xie C, Jiang XH, Zhang JT, Sun TT, Dong JD, Sanders AJ, Diao RY, Wang Y, Fok KL, Tsang LL, Yu MK, Zhang XH, Chung YW, Ye L, Zhao MY, Guo JH, Xiao ZJ, Lan HY, Ng CF, Lau KM, Cai ZM, Jiang WG, Chan HC. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32:2282-2291, 2291.e1-2291.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 177. | Zhang JT, Jiang XH, Xie C, Cheng H, Da Dong J, Wang Y, Fok KL, Zhang XH, Sun TT, Tsang LL, Chen H, Sun XJ, Chung YW, Cai ZM, Jiang WG, Chan HC. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochim Biophys Acta. 2013;1833:2961-2969. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 178. | Vij N, Mazur S, Zeitlin PL. CFTR is a negative regulator of NFkappaB mediated innate immune response. PLoS One. 2009;4:e4664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 137] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 179. | Massip-Copiz MM, Clauzure M, Valdivieso ÁG, Santa-Coloma TA. CFTR impairment upregulates c-Src activity through IL-1β autocrine signaling. Arch Biochem Biophys. 2017;616:1-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 180. | Liu K, Wang X, Zou C, Zhang J, Chen H, Tsang L, Yu MK, Chung YW, Wang J, Dai Y, Liu Y, Zhang X. Defective CFTR promotes intestinal proliferation via inhibition of the hedgehog pathway during cystic fibrosis. Cancer Lett. 2019;446:15-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 181. | Duranton C, Rubera I, Cougnon M, Melis N, Chargui A, Mograbi B, Tauc M. CFTR is involved in the fine tuning of intracellular redox status: physiological implications in cystic fibrosis. Am J Pathol. 2012;181:1367-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 182. | Fei Y, Sun L, Yuan C, Jiang M, Lou Q, Xu Y. CFTR ameliorates high glucose-induced oxidative stress and inflammation by mediating the NF-κB and MAPK signaling pathways in endothelial cells. Int J Mol Med. 2018;41:3501-3508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 183. | Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, Gavina M, Pulze L, Giardino I, Pettoello-Mantovani M, D'Apolito M, Guido S, Masliah E, Spencer B, Quaratino S, Raia V, Ballabio A, Maiuri L. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863-875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 372] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 184. | Zheng W, Kuhlicke J, Jäckel K, Eltzschig HK, Singh A, Sjöblom M, Riederer B, Weinhold C, Seidler U, Colgan SP, Karhausen J. Hypoxia inducible factor-1 (HIF-1)-mediated repression of cystic fibrosis transmembrane conductance regulator (CFTR) in the intestinal epithelium. FASEB J. 2009;23:204-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 185. | Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3:pii: e00116-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 186. | Birchenough GM, Nyström EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 315] [Cited by in F6Publishing: 362] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 187. | Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe. 2014;15:792-798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 184] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 188. | Li Y, Sun Z, Wu Y, Babovic-Vuksanovic D, Li Y, Cunningham JM, Pankratz VS, Yang P. Cystic fibrosis transmembrane conductance regulator gene mutation and lung cancer risk. Lung Cancer. 2010;70:14-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 189. | Yang X, Yan T, Gong Y, Liu X, Sun H, Xu W, Wang C, Naren D, Zheng Y. High CFTR expression in Philadelphia chromosome-positive acute leukemia protects and maintains continuous activation of BCR-ABL and related signaling pathways in combination with PP2A. Oncotarget. 2017;8:24437-24448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 190. | Peng X, Wu Z, Yu L, Li J, Xu W, Chan HC, Zhang Y, Hu L. Overexpression of cystic fibrosis transmembrane conductance regulator (CFTR) is associated with human cervical cancer malignancy, progression and prognosis. Gynecol Oncol. 2012;125:470-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 191. | Liu H, Wu W, Liu Y, Zhang C, Zhou Z. Predictive value of cystic fibrosis transmembrane conductance regulator (CFTR) in the diagnosis of gastric cancer. Clin Invest Med. 2014;37:E226-E232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 192. | Suh YS, Yu J, Kim BC, Choi B, Han TS, Ahn HS, Kong SH, Lee HJ, Kim WH, Yang HK. Overexpression of Plasminogen Activator Inhibitor-1 in Advanced Gastric Cancer with Aggressive Lymph Node Metastasis. Cancer Res Treat. 2015;47:718-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 193. | Li J, Zhang JT, Jiang X, Shi X, Shen J, Feng F, Chen J, Liu G, He P, Jiang J, Tsang LL, Wang Y, Rosell R, Jiang L, He J, Chan HC. The cystic fibrosis transmembrane conductance regulator as a biomarker in non-small cell lung cancer. Int J Oncol. 2015;46:2107-2115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 194. | Zhong X, Chen HQ, Yang XL, Wang Q, Chen W, Li C. CFTR activation suppresses glioblastoma cell proliferation, migration and invasion. Biochem Biophys Res Commun. 2019;508:1279-1285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 195. | Li W, Wang C, Peng X, Zhang H, Huang H, Liu H. CFTR inhibits the invasion and growth of esophageal cancer cells by inhibiting the expression of NF-κB. Cell Biol Int. 2018;42:1680-1687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 196. | Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, Buas MF, May A, Gerges C, Anders M, Becker J, Kreuser N, Noder T, Venerito M, Veits L, Schmidt T, Manner H, Schmidt C, Hess T, Böhmer AC, Izbicki JR, Hölscher AH, Lang H, Lorenz D, Schumacher B, Hackelsberger A, Mayershofer R, Pech O, Vashist Y, Ott K, Vieth M, Weismüller J, Nöthen MM; Barrett's and Esophageal Adenocarcinoma Consortium (BEACON); Esophageal Adenocarcinoma GenEtics Consortium (EAGLE); Wellcome Trust Case Control Consortium 2 (WTCCC2), Attwood S, Barr H, Chegwidden L, de Caestecker J, Harrison R, Love SB, MacDonald D, Moayyedi P, Prenen H, Watson RGP, Iyer PG, Anderson LA, Bernstein L, Chow WH, Hardie LJ, Lagergren J, Liu G, Risch HA, Wu AH, Ye W, Bird NC, Shaheen NJ, Gammon MD, Corley DA, Caldas C, Moebus S, Knapp M, Peters WHM, Neuhaus H, Rösch T, Ell C, MacGregor S, Pharoah P, Whiteman DC, Jankowski J, Schumacher J. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: A large-scale meta-analysis. Lancet Oncol. 2016;17:1363-1373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 197. | Cazacu IM, Farkas N, Garami A, Balaskó M, Mosdósi B, Alizadeh H, Gyöngyi Z, Rakonczay Z Jr, Vigh É, Habon T, Czopf L, Lazarescu MA, Erőss B, Sahin-Tóth M, Hegyi P. Pancreatitis-Associated Genes and Pancreatic Cancer Risk: A Systematic Review and Meta-analysis. Pancreas. 2018;47:1078-1086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 198. | McWilliams RR, Petersen GM, Rabe KG, Holtegaard LM, Lynch PJ, Bishop MD, Highsmith WE. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations and risk for pancreatic adenocarcinoma. Cancer. 2010;116:203-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 199. | Tu Z, Chen Q, Zhang JT, Jiang X, Xia Y, Chan HC. CFTR is a potential marker for nasopharyngeal carcinoma prognosis and metastasis. Oncotarget. 2016;7:76955-76965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 200. | Cystic Fibrosis Foundation. Carrier Testing for Cystic Fibrosis, 2019. Available from: https://www.cff.org. [Cited in This Article: ] |
| 201. | Shi J, Li H, Yuan C, Luo M, Wei J, Liu X. Cigarette Smoke-Induced Acquired Dysfunction of Cystic Fibrosis Transmembrane Conductance Regulator in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Oxid Med Cell Longev. 2018;2018:6567578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 202. | Li H, Ma N, Wang J, Wang Y, Yuan C, Wu J, Luo M, Yang J, Chen J, Shi J, Liu X. Nicotine Induces Progressive Properties of Lung Adenocarcinoma A549 Cells by Inhibiting Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Expression and Plasma Membrane Localization. Technol Cancer Res Treat. 2018;17:1533033818809984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 203. | Cystic Fibrosis Foundation. CFTR Modulator Therapies, 2019. Available from: https://www.cff.org. [Cited in This Article: ] |
| 204. | Bodewes F, Wilschanski M. CFTR Protein Function Modulation Therapy Is Finally Targeting Cystic Fibrosis-related Gastrointestinal Disease. J Pediatr Gastroenterol Nutr. 2018;66:372-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 205. | Ferrera L, Baroni D, Moran O. Lumacaftor-rescued F508del-CFTR has a modified bicarbonate permeability. J Cyst Fibros. 2019;pii:S1569-1993(18)30927-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 206. | Ooi CY, Syed SA, Rossi L, Garg M, Needham B, Avolio J, Young K, Surette MG, Gonska T. Impact of CFTR modulation with Ivacaftor on Gut Microbiota and Intestinal Inflammation. Sci Rep. 2018;8:17834. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 207. | Lin WY, Sohma Y, Hwang TC. Synergistic Potentiation of Cystic Fibrosis Transmembrane Conductance Regulator Gating by Two Chemically Distinct Potentiators, Ivacaftor (VX-770) and 5-Nitro-2-(3-Phenylpropylamino) Benzoate. Mol Pharmacol. 2016;90:275-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 208. | Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, Bronsveld I, van de Graaf EA, Nieuwenhuis EE, Houwen RH, Vleggaar FP, Escher JC, de Rijke YB, Majoor CJ, Heijerman HG, de Winter-de Groot KM, Clevers H, van der Ent CK, Beekman JM. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8:344ra84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 368] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 209. | Suh KS, Yuspa SH. Intracellular chloride channels: Critical mediators of cell viability and potential targets for cancer therapy. Curr Pharm Des. 2005;11:2753-2764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 210. | Winpenny JP, Marsey LL, Sexton DW. The CLCA gene family: Putative therapeutic target for respiratory diseases. Inflamm Allergy Drug Targets. 2009;8:146-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 211. | Yang B, Cao L, Liu B, McCaig CD, Pu J. The transition from proliferation to differentiation in colorectal cancer is regulated by the calcium activated chloride channel A1. PLoS One. 2013;8:e60861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 212. | Bustin SA, Li SR, Dorudi S. Expression of the Ca2+-activated chloride channel genes CLCA1 and CLCA2 is downregulated in human colorectal cancer. DNA Cell Biol. 2001;20:331-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 213. | Li X, Hu W, Zhou J, Huang Y, Peng J, Yuan Y, Yu J, Zheng S. CLCA1 suppresses colorectal cancer aggressiveness via inhibition of the Wnt/beta-catenin signaling pathway. Cell Commun Signal. 2017;15:38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 214. | Hu D, Ansari D, Zhou Q, Sasor A, Hilmersson KS, Bauden M, Jiang Y, Andersson R. Calcium-activated chloride channel regulator 1 as a prognostic biomarker in pancreatic ductal adenocarcinoma. BMC Cancer. 2018;18:1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 215. | Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, Valenzuela SM, Mazzanti M, Brown LJ, Breit SN, Curmi PM. The enigma of the CLIC proteins: Ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010;584:2093-2101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 216. | Petrova DT, Asif AR, Armstrong VW, Dimova I, Toshev S, Yaramov N, Oellerich M, Toncheva D. Expression of chloride intracellular channel protein 1 (CLIC1) and tumor protein D52 (TPD52) as potential biomarkers for colorectal cancer. Clin Biochem. 2008;41:1224-1236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 217. | Jia N, Dong S, Zhao G, Gao H, Li X, Zhang H. CLIC1 overexpression is associated with poor prognosis in pancreatic ductal adenocarcinomas. J Cancer Res Ther. 2016;12:892-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 218. | Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta. 2015;1848:2523-2531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 219. | Lu J, Dong Q, Zhang B, Wang X, Ye B, Zhang F, Song X, Gao G, Mu J, Wang Z, Ma F, Gu J. Chloride intracellular channel 1 (CLIC1) is activated and functions as an oncogene in pancreatic cancer. Med Oncol. 2015;32:616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 220. | Ma PF, Chen JQ, Wang Z, Liu JL, Li BP. Function of chloride intracellular channel 1 in gastric cancer cells. World J Gastroenterol. 2012;18:3070-3080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 35] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 221. | Chen CD, Wang CS, Huang YH, Chien KY, Liang Y, Chen WJ, Lin KH. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7:155-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 222. | Zhao W, Lu M, Zhang Q. Chloride intracellular channel 1 regulates migration and invasion in gastric cancer by triggering the ROS-mediated p38 MAPK signaling pathway. Mol Med Rep. 2015;12:8041-8047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 223. | Li BP, Mao YT, Wang Z, Chen YY, Wang Y, Zhai CY, Shi B, Liu SY, Liu JL, Chen JQ. CLIC1 Promotes the Progression of Gastric Cancer by Regulating the MAPK/AKT Pathways. Cell Physiol Biochem. 2018;46:907-924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 224. | Wei X, Li J, Xie H, Wang H, Wang J, Zhang X, Zhuang R, Lu D, Ling Q, Zhou L, Xu X, Zheng S. Chloride intracellular channel 1 participates in migration and invasion of hepatocellular carcinoma by targeting maspin. J Gastroenterol Hepatol. 2015;30:208-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 225. | Wang JW, Peng SY, Li JT, Wang Y, Zhang ZP, Cheng Y, Cheng DQ, Weng WH, Wu XS, Fei XZ, Quan ZW, Li JY, Li SG, Liu YB. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 226. | He YM, Zhang ZL, Liu QY, Xiao YS, Wei L, Xi C, Nan X. Effect of CLIC1 gene silencing on proliferation, migration, invasion and apoptosis of human gallbladder cancer cells. J Cell Mol Med. 2018;22:2569-2579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 227. | Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, McKay CJ, Carter R, Strachan D, Anderson K, Sansom OJ, Caswell PT, Norman JC. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22:131-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 242] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 228. | Hernandez-Fernaud JR, Ruengeler E, Casazza A, Neilson LJ, Pulleine E, Santi A, Ismail S, Lilla S, Dhayade S, MacPherson IR, McNeish I, Ennis D, Ali H, Kugeratski FG, Al Khamici H, van den Biggelaar M, van den Berghe PV, Cloix C, McDonald L, Millan D, Hoyle A, Kuchnio A, Carmeliet P, Valenzuela SM, Blyth K, Yin H, Mazzone M, Norman JC, Zanivan S. Secreted CLIC3 drives cancer progression through its glutathione-dependent oxidoreductase activity. Nat Commun. 2017;8:14206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 229. | Andersen AP, Moreira JM, Pedersen SF. Interactions of ion transporters and channels with cancer cell metabolism and the tumour microenvironment. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 230. | Deng YJ, Tang N, Liu C, Zhang JY, An SL, Peng YL, Ma LL, Li GQ, Jiang Q, Hu CT, Wang YN, Liang YZ, Bian XW, Fang WG, Ding YQ. CLIC4, ERp29, and Smac/DIABLO derived from metastatic cancer stem-like cells stratify prognostic risks of colorectal cancer. Clin Cancer Res. 2014;20:3809-3817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 231. | Zou Q, Yang Z, Li D, Liu Z, Yuan Y. Association of chloride intracellular channel 4 and Indian hedgehog proteins with survival of patients with pancreatic ductal adenocarcinoma. Int J Exp Pathol. 2016;97:422-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 232. | Sui Y, Sun M, Wu F, Yang L, Di W, Zhang G, Zhong L, Ma Z, Zheng J, Fang X, Ma T. Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells. PLoS One. 2014;9:e115443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 233. | Jiang Y, Cai Y, Shao W, Li F, Guan Z, Zhou Y, Tang C, Feng S. MicroRNA‑144 suppresses aggressive phenotypes of tumor cells by targeting ANO1 in colorectal cancer. Oncol Rep. 2019;41:2361-2370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 234. | Ji Q, Guo S, Wang X, Pang C, Zhan Y, Chen Y, An H. Recent advances in TMEM16A: Structure, function, and disease. J Cell Physiol. 2019;234:7856-7873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 235. | Park YR, Lee ST, Kim SL, Zhu SM, Lee MR, Kim SH, Kim IH, Lee SO, Seo SY, Kim SW. Down-regulation of miR-9 promotes epithelial mesenchymal transition via regulating anoctamin-1 (ANO1) in CRC cells. Cancer Genet. 2019;231-232:22-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 236. | Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, Mizushima T, Doki Y, Mori M, Yamamoto H. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann Surg Oncol. 2016;23:599-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 237. | Sauter DRP, Novak I, Pedersen SF, Larsen EH, Hoffmann EK. ANO1 (TMEM16A) in pancreatic ductal adenocarcinoma (PDAC). Pflugers Arch. 2015;467:1495-1508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 238. | Shang L, Hao JJ, Zhao XK, He JZ, Shi ZZ, Liu HJ, Wu LF, Jiang YY, Shi F, Yang H, Zhang Y, Liu YZ, Zhang TT, Xu X, Cai Y, Jia XM, Li M, Zhan QM, Li EM, Wang LD, Wei WQ, Wang MR. ANO1 protein as a potential biomarker for esophageal cancer prognosis and precancerous lesion development prediction. Oncotarget. 2016;7:24374-24382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 239. | Lu G, Shi W, Zheng H. Inhibition of STAT6/Anoctamin-1 Activation Suppresses Proliferation and Invasion of Gastric Cancer Cells. Cancer Biother Radiopharm. 2018;33:3-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 240. | Berglund E, Akcakaya P, Berglund D, Karlsson F, Vukojević V, Lee L, Bogdanović D, Lui WO, Larsson C, Zedenius J, Fröbom R, Bränström R. Functional role of the Ca²⁺-activated Cl⁻ channel DOG1/TMEM16A in gastrointestinal stromal tumor cells. Exp Cell Res. 2014;326:315-325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 241. | Simon S, Grabellus F, Ferrera L, Galietta L, Schwindenhammer B, Mühlenberg T, Taeger G, Eilers G, Treckmann J, Breitenbuecher F, Schuler M, Taguchi T, Fletcher JA, Bauer S. DOG1 regulates growth and IGFBP5 in gastrointestinal stromal tumors. Cancer Res. 2013;73:3661-3670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 242. | West RB, Corless CL, Chen X, Rubin BP, Subramanian S, Montgomery K, Zhu S, Ball CA, Nielsen TO, Patel R, Goldblum JR, Brown PO, Heinrich MC, van de Rijn M. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am J Pathol. 2004;165:107-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 462] [Cited by in F6Publishing: 440] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 243. | Fink AK, Yanik EL, Marshall BC, Wilschanski M, Lynch CF, Austin AA, Copeland G, Safaeian M, Engels EA. Cancer risk among lung transplant recipients with cystic fibrosis. J Cyst Fibros. 2017;16:91-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 244. | Ibele AR, Koplin SA, Slaughenhoupt BL, Kryger JV, Friedl A, Lund DP. Colonic adenocarcinoma in a 13-year-old with cystic fibrosis. J Pediatr Surg. 2007;42:E1-E3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 245. | Chaun H, Paty B, Nakielna EM, Schmidt N, Holden JK, Melosky B. Colonic carcinoma in two adult cystic fibrosis patients. Can J Gastroenterol. 1996;10:440-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 246. | McKenna PB, Mulcahy E, Waldron D. Early onset of colonic adenocarcinoma associated with cystic fibrosis--a case report. Ir Med J. 2006;99:310-311. [PubMed] [Cited in This Article: ] |
| 247. | Breuer W, Slotki IN, Ausiello DA, Cabantchik IZ. Induction of multidrug resistance downregulates the expression of CFTR in colon epithelial cells. Am J Physiol. 1993;265:C1711-C1715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 47] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 248. | McWilliams R, Highsmith WE, Rabe KG, de Andrade M, Tordsen LA, Holtegaard LM, Petersen GM. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut. 2005;54:1661-1662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 249. | Singh AP, Chauhan SC, Andrianifahanana M, Moniaux N, Meza JL, Copin MC, van Seuningen I, Hollingsworth MA, Aubert JP, Batra SK. MUC4 expression is regulated by cystic fibrosis transmembrane conductance regulator in pancreatic adenocarcinoma cells via transcriptional and post-translational mechanisms. Oncogene. 2007;26:30-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 250. | Hamoir C, Pepermans X, Piessevaux H, Jouret-Mourin A, Weynand B, Habyalimana JB, Leal T, Geubel A, Gigot JF, Deprez PH. Clinical and morphological characteristics of sporadic genetically determined pancreatitis as compared to idiopathic pancreatitis: Higher risk of pancreatic cancer in CFTR variants. Digestion. 2013;87:229-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 251. | Johannesson M, Askling J, Montgomery SM, Ekbom A, Bahmanyar S. Cancer risk among patients with cystic fibrosis and their first-degree relatives. Int J Cancer. 2009;125:2953-2956. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 252. | Moribe T, Iizuka N, Miura T, Kimura N, Tamatsukuri S, Ishitsuka H, Hamamoto Y, Sakamoto K, Tamesa T, Oka M. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer. 2009;125:388-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 253. | Mignen O, Constantin B, Potier-Cartereau M, Penna A, Gautier M, Guéguinou M, Renaudineau Y, Shoji KF, Félix R, Bayet E, Buscaglia P, Debant M, Chantôme A, Vandier C. Constitutive calcium entry and cancer: Updated views and insights. Eur Biophys J. 2017;46:395-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 254. | Xie J, Pan H, Yao J, Zhou Y, Han W. SOCE and cancer: Recent progress and new perspectives. Int J Cancer. 2016;138:2067-2077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 255. | Wang CY, Lai MD, Phan NN, Sun Z, Lin YC. Meta-Analysis of Public Microarray Datasets Reveals Voltage-Gated Calcium Gene Signatures in Clinical Cancer Patients. PLoS One. 2015;10:e0125766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 256. | Villalobos C, Sobradillo D, Hernández-Morales M, Núñez L. Calcium remodeling in colorectal cancer. Biochim Biophys Acta Mol Cell Res. 2017;1864:843-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 257. | Zhang Z, Liu X, Feng B, Liu N, Wu Q, Han Y, Nie Y, Wu K, Shi Y, Fan D. STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene. 2015;34:4808-4820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 258. | Shapovalov G, Ritaine A, Skryma R, Prevarskaya N. Role of TRP ion channels in cancer and tumorigenesis. Semin Immunopathol. 2016;38:357-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 259. | Yee NS, Chan AS, Yee JD, Yee RK. TRPM7 and TRPM8 Ion Channels in Pancreatic Adenocarcinoma: Potential Roles as Cancer Biomarkers and Targets. Scientifica (Cairo). 2012;2012:415158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 260. | Wong KK, Banham AH, Yaacob NS, Nur Husna SM. The oncogenic roles of TRPM ion channels in cancer. J Cell Physiol. 2019;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 261. | Hantute-Ghesquier A, Haustrate A, Prevarskaya N, Lehen'kyi V. TRPM Family Channels in Cancer. Pharmaceuticals (Basel). 2018;11:pii: E58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 262. | Yee NS, Zhou W, Lee M. Transient receptor potential channel TRPM8 is over-expressed and required for cellular proliferation in pancreatic adenocarcinoma. Cancer Lett. 2010;297:49-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 263. | Rybarczyk P, Gautier M, Hague F, Dhennin-Duthille I, Chatelain D, Kerr-Conte J, Pattou F, Regimbeau JM, Sevestre H, Ouadid-Ahidouch H. Transient receptor potential melastatin-related 7 channel is overexpressed in human pancreatic ductal adenocarcinomas and regulates human pancreatic cancer cell migration. Int J Cancer. 2012;131:E851-E861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 264. | Rybarczyk P, Vanlaeys A, Brassart B, Dhennin-Duthille I, Chatelain D, Sevestre H, Ouadid-Ahidouch H, Gautier M. The Transient Receptor Potential Melastatin 7 Channel Regulates Pancreatic Cancer Cell Invasion through the Hsp90α/uPA/MMP2 pathway. Neoplasia. 2017;19:288-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 265. | Yee NS, Kazi AA, Li Q, Yang Z, Berg A, Yee RK. Aberrant over-expression of TRPM7 ion channels in pancreatic cancer: Required for cancer cell invasion and implicated in tumor growth and metastasis. Biol Open. 2015;4:507-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 266. | Yee NS, Zhou W, Lee M, Yee RK. Targeted silencing of TRPM7 ion channel induces replicative senescence and produces enhanced cytotoxicity with gemcitabine in pancreatic adenocarcinoma. Cancer Lett. 2012;318:99-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 267. | Yee NS. Role of TRPM7 in Cancer: Potential as Molecular Biomarker and Therapeutic Target. Pharmaceuticals (Basel). 2017;10:pii: E39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 268. | Dai Q, Shrubsole MJ, Ness RM, Schlundt D, Cai Q, Smalley WE, Li M, Shyr Y, Zheng W. The relation of magnesium and calcium intakes and a genetic polymorphism in the magnesium transporter to colorectal neoplasia risk. Am J Clin Nutr. 2007;86:743-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 269. | Kim BJ, Park EJ, Lee JH, Jeon JH, Kim SJ, So I. Suppression of transient receptor potential melastatin 7 channel induces cell death in gastric cancer. Cancer Sci. 2008;99:2502-2509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 270. | Kim BJ, Kim SY, Lee S, Jeon JH, Matsui H, Kwon YK, Kim SJ, So I. The role of transient receptor potential channel blockers in human gastric cancer cell viability. Can J Physiol Pharmacol. 2012;90:175-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 271. | Kim BJ, Hong C. Role of transient receptor potential melastatin type 7 channel in gastric cancer. Integr Med Res. 2016;5:124-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 272. | Lin R, Wang Y, Chen Q, Liu Z, Xiao S, Wang B, Shi B. TRPM2 promotes the proliferation and invasion of pancreatic ductal adenocarcinoma. Mol Med Rep. 2018;17:7537-7544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 273. | Almasi S, Kennedy BE, El-Aghil M, Sterea AM, Gujar S, Partida-Sánchez S, El Hiani Y. TRPM2 channel-mediated regulation of autophagy maintains mitochondrial function and promotes gastric cancer cell survival via the JNK-signaling pathway. J Biol Chem. 2018;293:3637-3650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 274. | Sobradillo D, Hernández-Morales M, Ubierna D, Moyer MP, Núñez L, Villalobos C. A reciprocal shift in transient receptor potential channel 1 (TRPC1) and stromal interaction molecule 2 (STIM2) contributes to Ca2+ remodeling and cancer hallmarks in colorectal carcinoma cells. J Biol Chem. 2014;289:28765-28782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 275. | Zhu H, Zhang H, Jin F, Fang M, Huang M, Yang CS, Chen T, Fu L, Pan Z. Elevated Orai1 expression mediates tumor-promoting intracellular Ca2+ oscillations in human esophageal squamous cell carcinoma. Oncotarget. 2014;5:3455-3471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 114] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 276. | Shi Y, Ding X, He ZH, Zhou KC, Wang Q, Wang YZ. Critical role of TRPC6 channels in G2 phase transition and the development of human oesophageal cancer. Gut. 2009;58:1443-1450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 277. | Zhang SS, Wen J, Yang F, Cai XL, Yang H, Luo KJ, Liu QW, Hu RG, Xie X, Huang QY, Chen JY, Fu JH, Hu Y. High expression of transient potential receptor C6 correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Med Oncol. 2013;30:607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 278. | Cai R, Ding X, Zhou K, Shi Y, Ge R, Ren G, Jin Y, Wang Y. Blockade of TRPC6 channels induced G2/M phase arrest and suppressed growth in human gastric cancer cells. Int J Cancer. 2009;125:2281-2287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 279. | Kondratska K, Kondratskyi A, Yassine M, Lemonnier L, Lepage G, Morabito A, Skryma R, Prevarskaya N. Orai1 and STIM1 mediate SOCE and contribute to apoptotic resistance of pancreatic adenocarcinoma. Biochim Biophys Acta. 2014;1843:2263-2269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 280. | Crottès D, Rapetti-Mauss R, Alcaraz-Perez F, Tichet M, Gariano G, Martial S, Guizouarn H, Pellissier B, Loubat A, Popa A, Paquet A, Presta M, Tartare-Deckert S, Cayuela ML, Martin P, Borgese F, Soriani O. SIGMAR1 Regulates Membrane Electrical Activity in Response to Extracellular Matrix Stimulation to Drive Cancer Cell Invasiveness. Cancer Res. 2016;76:607-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 281. | Skrzycki M, Czeczot H. Altered expression level of Sigma1 receptor gene in human colorectal cancer. J Recept Signal Transduct Res. 2013;33:313-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 282. | Gueguinou M, Crottès D, Chantôme A, Rapetti-Mauss R, Potier-Cartereau M, Clarysse L, Girault A, Fourbon Y, Jézéquel P, Guérin-Charbonnel C, Fromont G, Martin P, Pellissier B, Schiappa R, Chamorey E, Mignen O, Uguen A, Borgese F, Vandier C, Soriani O. The SigmaR1 chaperone drives breast and colorectal cancer cell migration by tuning SK3-dependent Ca2+ homeostasis. Oncogene. 2017;36:3640-3647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 283. | Wang XT, Nagaba Y, Cross HS, Wrba F, Zhang L, Guggino SE. The mRNA of L-type calcium channel elevated in colon cancer: Protein distribution in normal and cancerous colon. Am J Pathol. 2000;157:1549-1562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 89] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 284. | Fornaro L, Vivaldi C, Lin D, Xue H, Falcone A, Wang Y, Crea F, Bootman MD. Prognostic relevance of a T-type calcium channels gene signature in solid tumours: A correlation ready for clinical validation. PLoS One. 2017;12:e0182818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 285. | Xie B, Zhao R, Bai B, Wu Y, Xu Y, Lu S, Fang Y, Wang Z, Maswikiti EP, Zhou X, Pan H, Han W. Identification of key tumorigenesis‑related genes and their microRNAs in colon cancer. Oncol Rep. 2018;40:3551-3560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 286. | Toyota M, Ho C, Ohe-Toyota M, Baylin SB, Issa JP. Inactivation of CACNA1G, a T-type calcium channel gene, by aberrant methylation of its 5' CpG island in human tumors. Cancer Res. 1999;59:4535-4541. [PubMed] [Cited in This Article: ] |
| 287. | Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835-1839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 288. | Yuasa Y, Nagasaki H, Akiyama Y, Hashimoto Y, Takizawa T, Kojima K, Kawano T, Sugihara K, Imai K, Nakachi K. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int J Cancer. 2009;124:2677-2682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 289. | Wanajo A, Sasaki A, Nagasaki H, Shimada S, Otsubo T, Owaki S, Shimizu Y, Eishi Y, Kojima K, Nakajima Y, Kawano T, Yuasa Y, Akiyama Y. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;135:580-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 290. | Patel F, Brackenbury WJ. Dual roles of voltage-gated sodium channels in development and cancer. Int J Dev Biol. 2015;59:357-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 291. | Winters JJ, Isom LL. Developmental and Regulatory Functions of Na(+) Channel Non-pore-forming β Subunits. Curr Top Membr. 2016;78:315-351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 292. | Patino GA, Isom LL. Electrophysiology and beyond: Multiple roles of Na+ channel β subunits in development and disease. Neurosci Lett. 2010;486:53-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 293. | Wang W, Takashima S, Segawa Y, Itoh M, Shi X, Hwang SK, Nabeshima K, Takeshita M, Hirose S. The developmental changes of Na(v)1.1 and Na(v)1.2 expression in the human hippocampus and temporal lobe. Brain Res. 2011;1389:61-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 294. | House CD, Vaske CJ, Schwartz AM, Obias V, Frank B, Luu T, Sarvazyan N, Irby R, Strausberg RL, Hales TG, Stuart JM, Lee NH. Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer Res. 2010;70:6957-6967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 295. | House CD, Wang BD, Ceniccola K, Williams R, Simaan M, Olender J, Patel V, Baptista-Hon DT, Annunziata CM, Gutkind JS, Hales TG, Lee NH. Voltage-gated Na+ Channel Activity Increases Colon Cancer Transcriptional Activity and Invasion Via Persistent MAPK Signaling. Sci Rep. 2015;5:11541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 296. | Peng J, Ou Q, Wu X, Zhang R, Zhao Q, Jiang W, Lu Z, Wan D, Pan Z, Fang Y. Expression of voltage-gated sodium channel Nav1.5 in non-metastatic colon cancer and its associations with estrogen receptor (ER)-β expression and clinical outcomes. Chin J Cancer. 2017;36:89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 297. | Xia J, Huang N, Huang H, Sun L, Dong S, Su J, Zhang J, Wang L, Lin L, Shi M, Bin J, Liao Y, Li N, Liao W. Voltage-gated sodium channel Nav 1.7 promotes gastric cancer progression through MACC1-mediated upregulation of NHE1. Int J Cancer. 2016;139:2553-2569. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 298. | Luo AJ, Tan J, He LY, Jiang XZ, Jiang ZQ, Zeng Q, Yao K, Xue J. Suppression of Tescalcin inhibits growth and metastasis in renal cell carcinoma via downregulating NHE1 and NF-kB signaling. Exp Mol Pathol. 2019;107:110-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 299. | Wang H, Long X, Wang D, Lou M, Zou D, Chen R, Nian W, Zhou Q. Increased expression of Na+/H+ exchanger isoform 1 predicts tumor aggressiveness and unfavorable prognosis in epithelial ovarian cancer. Oncol Lett. 2018;16:6713-6720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 300. | Flinck M, Kramer SH, Schnipper J, Andersen AP, Pedersen SF. The acid-base transport proteins NHE1 and NBCn1 regulate cell cycle progression in human breast cancer cells. Cell Cycle. 2018;17:1056-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 301. | Xie R, Wang H, Jin H, Wen G, Tuo B, Xu J. NHE1 is upregulated in gastric cancer and regulates gastric cancer cell proliferation, migration and invasion. Oncol Rep. 2017;37:1451-1460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 302. | Benhaim L, Gerger A, Bohanes P, Paez D, Wakatsuki T, Yang D, Labonte MJ, Ning Y, El-Khoueiry R, Loupakis F, Zhang W, Laurent-Puig P, Lenz HJ. Gender-specific profiling in SCN1A polymorphisms and time-to-recurrence in patients with stage II/III colorectal cancer treated with adjuvant 5-fluoruracil chemotherapy. Pharmacogenomics J. 2014;14:135-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 303. | Igci YZ, Bozgeyik E, Borazan E, Pala E, Suner A, Ulasli M, Gurses SA, Yumrutas O, Balik AA, Igci M. Expression profiling of SCN8A and NDUFC2 genes in colorectal carcinoma. Exp Oncol. 2015;37:77-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 304. | Driffort V, Gillet L, Bon E, Marionneau-Lambot S, Oullier T, Joulin V, Collin C, Pagès JC, Jourdan ML, Chevalier S, Bougnoux P, Le Guennec JY, Besson P, Roger S. Ranolazine inhibits NaV1.5-mediated breast cancer cell invasiveness and lung colonization. Mol Cancer. 2014;13:264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 305. | Nelson M, Yang M, Millican-Slater R, Brackenbury WJ. Nav1.5 regulates breast tumor growth and metastatic dissemination in vivo. Oncotarget. 2015;6:32914-32929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 306. | Levaot N, Hershfinkel M. How cellular Zn2+ signaling drives physiological functions. Cell Calcium. 2018;75:53-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 307. | Hershfinkel M. The Zinc Sensing Receptor, ZnR/GPR39, in Health and Disease. Int J Mol Sci. 2018;19:pii: E439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 308. | Pan Z, Choi S, Ouadid-Ahidouch H, Yang JM, Beattie JH, Korichneva I. Zinc transporters and dysregulated channels in cancers. Front Biosci (Landmark Ed). 2017;22:623-643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 309. | Inoue K, O'Bryant Z, Xiong ZG. Zinc-permeable ion channels: Effects on intracellular zinc dynamics and potential physiological/pathophysiological significance. Curr Med Chem. 2015;22:1248-1257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 310. | Schweigel-Röntgen M. The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr Top Membr. 2014;73:321-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 311. | Fukada T, Kambe T. Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics. 2011;3:662-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 312. | Ohashi W, Hara T, Takagishi T, Hase K, Fukada T. Maintenance of Intestinal Epithelial Homeostasis by Zinc Transporters. Dig Dis Sci. 2019;64:2404-2415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 313. | Yang J, Zhang Y, Cui X, Yao W, Yu X, Cen P, Hodges SE, Fisher WE, Brunicardi FC, Chen C, Yao Q, Li M. Gene profile identifies zinc transporters differentially expressed in normal human organs and human pancreatic cancer. Curr Mol Med. 2013;13:401-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 314. | Semrad CE. Zinc and intestinal function. Curr Gastroenterol Rep. 1999;1:398-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 315. | Myers S, Shastri MD, Adulcikas J, Sohal SS, Norouzi S. Zinc and Gastrointestinal Disorders: A Role for the Zinc Transporters Zips and ZnTs. Curr Pharm Des. 2017;23:2328-2332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 316. | Goh J, O'Morain CA. Review article: Nutrition and adult inflammatory bowel disease. Aliment Pharmacol Ther. 2003;17:307-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 317. | Hering NA, Schulzke JD. Therapeutic options to modulate barrier defects in inflammatory bowel disease. Dig Dis. 2009;27:450-454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 318. | Cohen L, Azriel-Tamir H, Arotsker N, Sekler I, Hershfinkel M. Zinc sensing receptor signaling, mediated by GPR39, reduces butyrate-induced cell death in HT29 colonocytes via upregulation of clusterin. PLoS One. 2012;7:e35482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 319. | Cohen L, Sekler I, Hershfinkel M. The zinc sensing receptor, ZnR/GPR39, controls proliferation and differentiation of colonocytes and thereby tight junction formation in the colon. Cell Death Dis. 2014;5:e1307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 320. | Pongkorpsakol P, Buasakdi C, Chantivas T, Chatsudthipong V, Muanprasat C. An agonist of a zinc-sensing receptor GPR39 enhances tight junction assembly in intestinal epithelial cells via an AMPK-dependent mechanism. Eur J Pharmacol. 2019;842:306-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 321. | Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. J Biol Chem. 2006;281:24085-24089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 533] [Cited by in F6Publishing: 468] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 322. | Bafaro E, Liu Y, Xu Y, Dempski RE. The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct Target Ther. 2017;2:pii: 17029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 323. | Bhutia YD, Babu E, Ramachandran S, Yang S, Thangaraju M, Ganapathy V. SLC transporters as a novel class of tumour suppressors: Identity, function and molecular mechanisms. Biochem J. 2016;473:1113-1124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 324. | Takatani-Nakase T. Zinc Transporters and the Progression of Breast Cancers. Biol Pharm Bull. 2018;41:1517-1522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 325. | Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang SM, Cousins RJ, Fisher WE, Brunicardi FC, Logsdon CD, Chen C, Yao Q. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci U S A. 2007;104:18636-18641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 194] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 326. | Li M, Zhang Y, Bharadwaj U, Zhai QJ, Ahern CH, Fisher WE, Brunicardi FC, Logsdon CD, Chen C, Yao Q. Down-regulation of ZIP4 by RNA interference inhibits pancreatic cancer growth and increases the survival of nude mice with pancreatic cancer xenografts. Clin Cancer Res. 2009;15:5993-6001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 327. | Jin H, Liu P, Wu Y, Meng X, Wu M, Han J, Tan X. Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 2018;109:2946-2956. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 328. | Zhang Y, Yang J, Cui X, Chen Y, Zhu VF, Hagan JP, Wang H, Yu X, Hodges SE, Fang J, Chiao PJ, Logsdon CD, Fisher WE, Brunicardi FC, Chen C, Yao Q, Fernandez-Zapico ME, Li M. A novel epigenetic CREB-miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO Mol Med. 2013;5:1322-1334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 329. | Xu C, Wallace MB, Yang J, Jiang L, Zhai Q, Zhang Y, Hong C, Chen Y, Frank TS, Stauffer JA, Asbun HJ, Raimondo M, Woodward TA, Li Z, Guha S, Zheng L, Li M. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: A systemic comparison between EUS-FNA and surgical specimens. Curr Mol Med. 2014;14:309-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 330. | Unno J, Satoh K, Hirota M, Kanno A, Hamada S, Ito H, Masamune A, Tsukamoto N, Motoi F, Egawa S, Unno M, Horii A, Shimosegawa T. LIV-1 enhances the aggressive phenotype through the induction of epithelial to mesenchymal transition in human pancreatic carcinoma cells. Int J Oncol. 2009;35:813-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 331. | Costello LC, Levy BA, Desouki MM, Zou J, Bagasra O, Johnson LA, Hanna N, Franklin RB. Decreased zinc and downregulation of ZIP3 zinc uptake transporter in the development of pancreatic adenocarcinoma. Cancer Biol Ther. 2011;12:297-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 332. | Costello LC, Zou J, Desouki MM, Franklin RB. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. J Gastrointest Cancer. 2012;43:570-578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 333. | Wu C, Li D, Jia W, Hu Z, Zhou Y, Yu D, Tong T, Wang M, Lin D, Qiao Y, Zhou Y, Chang J, Zhai K, Wang M, Wei L, Tan W, Shen H, Zeng Y, Lin D. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet. 2013;45:632-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 334. | Jin J, Li Z, Liu J, Wu Y, Gao X, He Y. Knockdown of zinc transporter ZIP5 (SLC39A5) expression significantly inhibits human esophageal cancer progression. Oncol Rep. 2015;34:1431-1439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 335. | Kumar A, Chatopadhyay T, Raziuddin M, Ralhan R. Discovery of deregulation of zinc homeostasis and its associated genes in esophageal squamous cell carcinoma using cDNA microarray. Int J Cancer. 2007;120:230-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 336. | Choi S, Cui C, Luo Y, Kim SH, Ko JK, Huo X, Ma J, Fu LW, Souza RF, Korichneva I, Pan Z. Selective inhibitory effects of zinc on cell proliferation in esophageal squamous cell carcinoma through Orai1. FASEB J. 2018;32:404-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 337. | Barresi V, Valenti G, Spampinato G, Musso N, Castorina S, Rizzarelli E, Condorelli DF. Transcriptome analysis reveals an altered expression profile of zinc transporters in colorectal cancer. J Cell Biochem. 2018;119:9707-9719. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 338. | Ding B, Lou W, Xu L, Li R, Fan W. Analysis the prognostic values of solute carrier (SLC) family 39 genes in gastric cancer. Am J Transl Res. 2019;11:486-498. [PubMed] [Cited in This Article: ] |
| 339. | Sheng N, Yan L, You W, Tan G, Gong J, Chen H, Yang Y, Hu L, Wang Z. Knockdown of SLC39A7 inhibits cell growth and induces apoptosis in human colorectal cancer cells. Acta Biochim Biophys Sin (Shanghai). 2017;49:926-934. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 340. | Ohashi W, Kimura S, Iwanaga T, Furusawa Y, Irié T, Izumi H, Watanabe T, Hijikata A, Hara T, Ohara O, Koseki H, Sato T, Robine S, Mori H, Hattori Y, Watarai H, Mishima K, Ohno H, Hase K, Fukada T. Zinc Transporter SLC39A7/ZIP7 Promotes Intestinal Epithelial Self-Renewal by Resolving ER Stress. PLoS Genet. 2016;12:e1006349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 341. | Myers SA. Zinc transporters and zinc signaling: New insights into their role in type 2 diabetes. Int J Endocrinol. 2015;2015:167503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 342. | Bin BH, Seo J, Kim ST. Function, Structure, and Transport Aspects of ZIP and ZnT Zinc Transporters in Immune Cells. J Immunol Res. 2018;2018:9365747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 343. | Franklin RB, Zou J, Costello LC. The cytotoxic role of RREB1, ZIP3 zinc transporter, and zinc in human pancreatic adenocarcinoma. Cancer Biol Ther. 2014;15:1431-1437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 344. | Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, Andrews GK. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS One. 2010;5:pii: e13158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 345. | Xu X, Guo HJ, Xie HY, Li J, Zhuang RZ, Ling Q, Zhou L, Wei XY, Liu ZK, Ding SM, Chen KJ, Xu ZY, Zheng SS. ZIP4, a novel determinant of tumor invasion in hepatocellular carcinoma, contributes to tumor recurrence after liver transplantation. Int J Biol Sci. 2014;10:245-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 346. | Gartmann L, Wex T, Grüngreiff K, Reinhold D, Kalinski T, Malfertheiner P, Schütte K. Expression of zinc transporters ZIP4, ZIP14 and ZnT9 in hepatic carcinogenesis-An immunohistochemical study. J Trace Elem Med Biol. 2018;49:35-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 347. | Shen R, Xie F, Shen H, liu Q, Zheng T, Kou X, Wang D, Yang J. Negative correlation of LIV-1 and E-cadherin expression in hepatocellular carcinoma cells. PLoS One. 2013;8:e56542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 348. | Franklin RB, Levy BA, Zou J, Hanna N, Desouki MM, Bagasra O, Johnson LA, Costello LC. ZIP14 zinc transporter downregulation and zinc depletion in the development and progression of hepatocellular cancer. J Gastrointest Cancer. 2012;43:249-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 349. | Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, Zmoos AF, Vaka D, Tran KQ, Zhou M, Krasinska K, Riess JW, Neal JW, Khatri P, Park KS, Butte AJ, Sage J. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364-1377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 251] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 350. | Kale VP, Amin SG, Pandey MK. Targeting ion channels for cancer therapy by repurposing the approved drugs. Biochim Biophys Acta. 2015;1848:2747-2755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 351. | Humphries ES, Dart C. Neuronal and Cardiovascular Potassium Channels as Therapeutic Drug Targets: Promise and Pitfalls. J Biomol Screen. 2015;20:1055-1073. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |