Peer-review started: May 3, 2015
First decision: August 4, 2015
Revised: October 18, 2015
Accepted: November 10, 2015
Article in press: November 11, 2015
Published online: January 26, 2016
Atherosclerotic coronary artery disease (CAD) comprises a broad spectrum of clinical entities that include asymptomatic subclinical atherosclerosis and its clinical complications, such as angina pectoris, myocardial infarction (MI) and sudden cardiac death. CAD continues to be the leading cause of death in industrialized society. The long-recognized familial clustering of CAD suggests that genetics plays a central role in its development, with the heritability of CAD and MI estimated at approximately 50% to 60%. Understanding the genetic architecture of CAD and MI has proven to be difficult and costly due to the heterogeneity of clinical CAD and the underlying multi-decade complex pathophysiological processes that involve both genetic and environmental interactions. This review describes the clinical heterogeneity of CAD and MI to clarify the disease spectrum in genetic studies, provides a brief overview of the historical understanding and estimation of the heritability of CAD and MI, recounts major gene discoveries of potential causal mutations in familial CAD and MI, summarizes CAD and MI-associated genetic variants identified using candidate gene approaches and genome-wide association studies (GWAS), and summarizes the current status of the construction and validations of genetic risk scores for lifetime risk prediction and guidance for preventive strategies. Potential protective genetic factors against the development of CAD and MI are also discussed. Finally, GWAS have identified multiple genetic factors associated with an increased risk of in-stent restenosis following stent placement for obstructive CAD. This review will also address genetic factors associated with in-stent restenosis, which may ultimately guide clinical decision-making regarding revascularization strategies for patients with CAD and MI.
Core tip: This review provides the most comprehensive summary of the genetics of coronary artery disease (CAD) and myocardial infarction (MI) research with a complete, up-to-date chromosomal map of all CAD and MI-susceptible genes. We discuss the existence and significance of protective genetic factors against atherosclerosis, CAD and MI. We also summarize the current status of constructing genetic risk scores to predict long-term risks of developing CAD and MI. In-stent restenosis is a new challenge in cardiology. The genetics of in-stent restenosis are also discussed in this article.
- Citation: Dai X, Wiernek S, Evans JP, Runge MS. Genetics of coronary artery disease and myocardial infarction. World J Cardiol 2016; 8(1): 1-23
- URL: https://www.wjgnet.com/1949-8462/full/v8/i1/1.htm
- DOI: https://dx.doi.org/10.4330/wjc.v8.i1.1
Coronary artery disease (CAD) remains the number one cause of death in industrialized society. CAD alone caused approximately 1 of every 6 deaths in the United States in 2010[1]. Atherosclerotic CAD comprises a broad spectrum of clinical entities that include asymptomatic subclinical atherosclerosis and its clinical complications, such as angina pectoris, myocardial infarction (MI) and sudden cardiac death. In the early 1930s, Carl Miller in Oslo reported the co-segregation of high plasma cholesterol, xanthoma and premature coronary heart disease, providing early clues regarding a genetic component of CAD and its association with cholesterol[2]. Family clustering of CAD and MI has subsequently been well recognized and documented. Large twin studies have estimated the heritability of CAD to be approximately 50% to 60%. Understanding the genetic basis of CAD and MI will not only provide insight regarding the pathogenesis of the disease but also a basis for the development of preventive and therapeutic strategies. Research investigating the genetic architecture of CAD has proven to be a difficult and costly task due to the heterogeneities of clinical CAD and MI and its multi-decade complex pathophysiological processes that involve both genetics and environmental factors and their interactions. Together with rapid advances in molecular technology and computational capacities in genetics and genomics, the recent decade has witnessed tremendous progress in genetic and genomic studies of CAD and MI. This article provides a comprehensive overview of the clinical heterogeneity of CAD and MI, which are often assessed in genetic studies; the heritability of CAD and MI; and achievements in gene discoveries related to CAD, MI, and in-stent restenosis. The development of a genetic risk score (GRS) based on genetic risk factors related to CAD and its initial success for predicting the life-long risk of CAD is also discussed.
The heterogeneity of CAD and its clinical complications introduce significant complexity in genetic studies. Clinically, the presentation of atherosclerotic CAD ranges from completely asymptomatic (subclinical atherosclerosis), angina pectoris (typical or atypical, stable or unstable), and silent MI to acute myocardial infarction (AMI) or sudden cardiac death. The Framingham Heart Study (FHS) reported that one third of all MI are unrecognized[3]. Based on autopsy, up to 50% of sudden deaths are due to MI[4]. In addition to the broad spectrum of clinical presentations, the age of onset of clinical symptoms varies dramatically. The average age at the time of first MI is 64.9 years for men and 72.3 years for women[1]. Although AMI is not uncommon in young adults (< 40 years old), an initial diagnosis of severe atherosclerotic CAD in octogenarians is rather common and is often preceded by a long, asymptomatic disease state.
Coronary artery atherosclerosis is the most common underlying pathological process responsible for the majority of clinically significant CAD. Progressive narrowing of the arterial lumen due to negative remodeling and expansion of the atheroma causes myocardial ischemia and angina pectoris. The rupture of a vulnerable atherosclerotic plaque, local activation of thrombotic mechanisms with/without severe underlying stenosis, local thrombosis formation and arterial lumen closure are accepted as underlying mechanisms of AMI. Coronary embolization of the thrombus, spontaneous coronary dissection, myocardial bridging, an anomalous origin and course of coronary artery, and coronary spasm can cause clinical symptoms/presentations that are similar to those of AMI. In addition, some unusual clinical scenarios may cause coronary insufficiency and myocardial ischemia, such as adjacent tumor compression of coronary arteries and systemic vascularitis involving coronary artery beds. In addition, the locations of atherosclerotic stenosis along the coronary tree vary significantly. Isolated aorto-ostial stenosis (ostia of the left main or right coronary artery) and bifurcation lesions are more apparent in relation to turbulent flow and the endothelial response to the flow dynamics. Diffuse atherosclerosis is more commonly observed in patients with diabetes mellitus (DM). The heterogeneity of the location of CAD within the coronary tree may reflect a different set of genetic influences on atherogenesis. The location-dependent effects of PECAM-1 on atherogenesis in animal models have been reported[5], and the results suggest that the heterogeneity of the location of atherosclerotic CAD may represent different genetic influences of atherosclerosis under different dynamic flow conditions.
These heterogeneities of phenotypic characterization, pathological etiologies of CAD and MI and the complex molecular and cellular pathogenesis of atherosclerosis (summarized in Table 1) contribute to the difficulties associated with the identification of genes that are important for CAD and MI[6].
| Clinical manifestation | Underlying pathology | Pathological processes of atherosclerosis |
| Asymptomatic stenosis Stable or unstable angina pectoris Silent MI Acute MI (NSTEMI and STEMI) Sudden cardiac death | Atheroma positive remodeling Atheroma negative remodeling Plaque rupture/thrombosis Critical stenosis/thrombosis Embolization Spontaneous dissection Anomalous origin/course Coronary spasm Myocardial bridging | Endothelial injury Lipid deposition Oxidative stress/response Inflammation Cellular proliferation/apoptosis Foam cell formation Matrix deposition/degradation Plaque rupture/hematoma/thrombosis Neovascular formation |
Clinical and population-based studies have demonstrated that genetic factors play important roles in CAD and MI. The phenomenon of family clustering of CAD was repeatedly reported in the 1950s and 1960s[7,8]. Slack and Evans demonstrated that a history of early onset ischemic heart disease (IHD) of first degree relatives was significantly associated with and predicted early onset IHD (< 55 in men and < 65 in women)[9]. The subsequent FHS confirmed that a family history of premature CAD, defined as the presence of a first degree relative with a diagnosis of CAD at < 55 years of age in men and < 65 years of age in women, is an independent risk factor for CAD[10]. The strength of hereditary in determining the risk of CAD increases with an increasing number of affected first-degree relatives and with a younger age of onset[11]. In a multivariate analysis, the extent of CAD based on coronary angiography was also strongly associated with the history of parental CAD independently of plasma lipids, obesity, hypertension, cigarette smoking and alcohol intake[12-15].
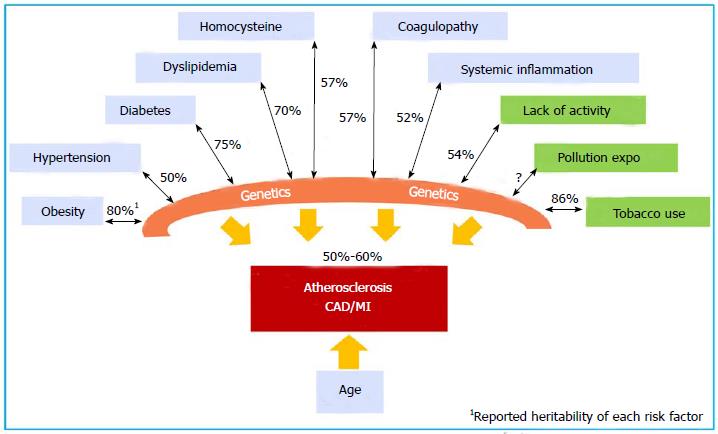
Twins have fascinated human communities and provided invaluable opportunities to identify the genetic component of diseases. Identical twins who concordantly develop early onset, angiographically proven CAD, in whom many of the same coronary arteries are even involved, provide highly suggestive information regarding the influence of genetics on CAD[16-20]. Early in 1967, Cederlöf et al[21] observed a concordance of 21.7% of CHD history in monozygotic twins, as compared with 6.1% in dizygotic twins. The Swedish Twin Registry and Danish Twins Registry are the two largest twin registries in the world. The Swedish Twin Registry has captured all of the twins born since 1886 and includes 20966 twins. A longitudinal follow-up of 36 years found that if one twin died of CAD, the relative risk of the development of fatal CAD in the second twin was 8.1 for monozygotic twins (MZ, identical twin)[22] and 3.8 for dizygotic twins (DZ, non-identical twin). The estimated heritability of CAD is 57% (50%-59%) in male twins and 38% (25%-50%) in female twins[23]. The influence of genetics is evident across the age range of 36 to 86 years. The Danish Twins Registry (8000 twin pairs) reported an increased incidence of CAD and CAD deaths in MZ twins of subjects with CAD compared with DZ twins (44% vs 14%), with an estimated heritability of 0.53 in males and 0.58 in females[24]. In general, CAD is widely accepted to have a heritability of 50% to 60%.
Large-scale prospective, population-based epidemiology studies, such as the Western Collaborative Group Study[25], Health Professional follow-up study[26], the Nurses’ Health Study[27], the FHS[10], the British Regional Heart Study[28], the German PROCAM study[29], and the Utah Cardiovascular Genetic Research[30], confirmed the strong independent association among family, parental history of CAD and MI and the occurrence in offspring[31,32]. These genetic factors are independent of traditional risk factors (TRF) for the disease. TRFs, such as hypertension, diabetes mellitus, hypercholesterolemia, low physical activity, obesity, C-reactive protein, plasma homocysteine, and tobacco use are well known to have their own complex genetic components with individual heritability values that have been estimated in twin studies[33]. Figure 1 illustrates the contribution of genetic factors directly or through TRFs in the development and manifestation of CAD. The collective effects of genetic factors, environmental factors, and age determine the development of atherosclerotic CAD and its complications.
Early candidate gene and linkage analyses have identified numerous causal genes and mutations that underlie rare, Mendelian monogenic CAD. Many of these genes and mutations are involved in lipid metabolism. Recently, a combination of a large pedigree of familial CAD and high-throughput genomic sequencing technology led to the discovery of a panel of new possible causal gene mutations for CAD. Table 2 summarizes the genes and mutations that are considered to be causal for atherosclerotic CAD.
| Categories | Genes | Chrom | OMIM | Mutations | Ref. | GWAS1 |
| Monogenic CAD genes | ST6GALNAC5 | 1p31.1 | 610134 | G295A (p.*337Qext*20 stop-loss) | [52] | No |
| CYP27A1 | 2p35 | 606530 | G674A (p. Arg225His) | [51] | No | |
| MEF2A | 15q26.3 | 608320 | 21-bp del in exon 11 | [34,38] | No | |
| LRP6 | 12p13.2 | 610947 | G1079A (p. Arg611Cys) T1298C (p. Asn433Ser) | [44,159] | No | |
| Gene mutations cause high LDL | LDL receptor | 19p13.2 | 606945 | > 1000 variants | [55] | Yes |
| PCSK9 | 1p32.3 | 603776 | 9 gain-of-function mutations | [63] | Yes | |
| ApoB-100 | 2p24.1 | 144010 | C10580G (p. Arg3527Gln) C10800T (p. Arg3531Cys) rs515135 | [59,60,84] | Yes | |
| LDLRAP1, ARH | 1p36.11 | 603813 | ARH1: 432 ins A (p. FS170stop) ARH2: G65A (p. Trp22ter) | [66] | No | |
| Mutations cause low HDL | ABCA1 | 9q31.1 | 205400 | Many ABCA1 LoF alleles Rs2230806 > A | [78,158] | Yes |
| LCAT | 16q22.1 | 606967 | > 80 mutations Rs5923 ↑ CAD in Egyptians | [160] | Yes | |
| Mutations cause high TG | Apo C-II | 19q13.2 | 207750 | ApoCIISt. Michael p. Gln70Pro | [85,86] | No |
Despite the clinical heterogeneity of CAD and MI as described above, the phenomenon of familial clustering of CAD and collections of large pedigrees with multiple members in multiple generations provided an opportunity to perform linkage analysis and gene discovery. In recent years, three potential causal genes and their responsible mutations for pedigrees with apparent Mendelian autosomal dominant (AD) CAD and MI have been identified (below and Table 2). The common feature of these mutations is co-segregation with the phenotype in the index kindred and a presence in unrelated cohorts. Functional analyses of these genes also supported their potential involvement in the pathogenesis of CAD and MI. However, the potential roles of these familial CAD causal genes and mutations in general population are unknown. Further validation with additional pedigrees and experimental models is warranted.
MEF2A: MEF2A is a transcription factor that belongs to the monocyte enhancer factor (MEF) family. MEF2A is expressed in blood vessels during embryogenesis as an early marker of vasculogenesis and interacts with a variety of molecules that are known to be involved in cardiovascular pathogenesis. Wang et al[34] performed a genome-wide linkage analysis of a large kindred group consisting of 13 individuals and designated the first AD CAD gene (adCAD1) at chromosomal location 15q26 within a region containing approximately 93 genes including MEF2A. Considering the relevance of MEF2A in vasculogenesis, sequencing of the MEF2A gene revealed a 21-base pair deletion in exon 11 of the MEF2A gene that was present in all 10 affected living members but not in unaffected individuals[34]. This D7aa MEF2A mutation co-segregated with the CAD phenotype in an AD manner in the index pedigree. Interestingly, the same exon 11 deletion in MEF2A was reported in other CAD individuals from different ethnic backgrounds[35-37]. Missense mutations in exon 7 of MEF2A, resulting in a loss-of-function of MEF2A, were also found in premature CAD individuals, but not in age-, ethnicity- and BMI-matched controls lacking angiographic evidence of CAD[38,39]. Further studies have demonstrated that the deletion of 7 amino acid leads to defective trafficking of MEF2A in a dominant-negative manner. Consequently, MEF2A entry into the nucleus is blocked, which is crucial for the ability of MEF2A to regulate gene expression. It is plausible that functional abnormalities in D7aa MEF2A lead to cellular abnormalities in endothelial cells and vascular smooth muscle cells, which in turn participate in processes associated with atherogenesis. The discovery of the mutation in MEF2A in CAD resulted in significant hope of genetic testing for CAD. However, conflicting reports demonstrating the lack of an association between MEF2A gene mutations and CAD in other cohorts has raised doubts concerning a causal role of MEF2A in CAD[40-42]. In particular, two individuals carrying the D7aa MEF2A mutation did not appear to have CAD before the reported age of CAD development, while members of the same family who developed CAD carried the normal MEF2A gene[41]. However, potential genetic or environmental modifiers may reduce the phenotypic penetration. Polymorphisms that do not change MEF2A transcriptional activation are not associated with an increased risk of CAD[43], and the actual prevalence of functional MEF2A mutations in the general population is not yet known.
Low-density lipoprotein receptor-related protein 6: Mani et al[44] studied a group of extreme outlier kindred with an extraordinary prevalence of premature CAD presenting an almost uniform presence of hypertension, hypercholesterolemia, type II DM, obesity and an absence of cigarette smoking in affected individuals. The extreme familial clustering and segregation of phenotypes (both premature CAD and risk factors) was transmitted as a highly penetrant AD trait[44]. Genome-wide linkage analysis revealed strong linkage between the familial trait and markers of chromosome 12p, which spans 750 kb and contains only six annotated genes [ETV6, BCL2L14, low-density lipoprotein receptor-related protein 6 (LRP6), MANSC1, LOH12CR1 and DUSP16]. Sequencing of the candidate gene LRP6 revealed the causal variant, a missense substitution of R611C in a conserved EGF-like domain of LRP6. LPR6 functions as a co-receptor with Frizzle proteins for Wnt ligands. Functional studies of the LRP6R611C mutation revealed a dominant-negative decrease in Wnt signaling. Although there was complete linkage of LRP6R611C and hypertension, high low-density lipoprotein (LDL), TG and a prevalence of DM2 in the index kindred, the frequency of LRP6R611C was very rare in general population. Recently, a polymorphism in LRP6 intron 2 was found to be associated with the presence and severity of angiographic CAD in a Chinese case-control study[45]. A common variant of LRP6, rs2302685 (a non-synonymous coding sequence SNP in exon 14, T>C, p Ile1062Val), was initially found to be associated with late-onset Alzheimer’s disease in Caucasians[46] followed by carotid artery atherosclerosis[47] and, very recently, CAD[48]. Ilr1062Val variant reduces Wnt/β-catenin signaling and anti-apoptotic activities in cultured cells and arterial walls. Additional missense mutations in the LRP6 gene that correlate to its extracellular domain have been identified by the sequencing of exons and the promoter of LRP6 in premature CAD patients and controls in different Chinese cohorts. All of these missense mutations cause loss of function (LoF) via reduced Wnt signaling activity and attenuated human umbilical vein endothelial cell proliferation in vitro[48].
CYP27A1: A large pedigree with well-defined familial traits together with massive parallel sequencing technology and bioinformatics computational and statistical tools has dramatically accelerated the pace of the discovery of disease-causing genes[49]. The identification of potential causative mutations in the CYP27A1 gene that co-segregated with a familial AD CAD phenotype is one recent example. Inanloorahatloo et al[50] performed whole-exome sequencing of two affected members in a large group of AD kindred with premature CAD. An in silico un-biased algorithm identified two candidate variants, c. G674A (p. Arg225His) in CYP27A1 and c. A241T (p.Ile81Phe) in NTRK2, which were further sequenced in all of the available members of the kindred. The variant c.G674A (p. Arg225His) in CYP27A1 co-segregated with the CAD status. The CYP27A1 gene encodes the sterol 27-hydroxylase involved in cholesterol and 25-hydroxy vitamin D3 synthesis. The amino acid p. Arg 225 that was affected by the identified variant is highly conserved in paralogous and orthologous proteins, suggesting its functional importance. This variant was not observed in 500 ethnically matched controls without a history of cardiac disease. Furthermore, an additional four disease-specific variants in the CYP27A1 gene were discovered by sequencing the CYP27A1 exons in 7 out of 100 unrelated CAD patients. Although disease-causing variants of the CYP27A1 gene were considered relatively rare, potential disease-causing variants reached up to 7% in Iranian patients with CAD. To date, potential CAD-causing CYP27A1 variants have not been reported in other populations, and the prevalence of these variants in the general population is unknown. The mechanism underlying the possible causal role of mutant CYP27A1 in atherosclerotic CAD remains uncharacterized. Recently, CYP27A1-deficient mice in an ApoE-deficient background exhibited a 10-fold reduction of aortic atherosclerosis after challenge with a high-fat diet associated with a 2-fold reduction of total plasma cholesterol, LDL, and very low-density lipoprotein (VLDL) as well as a 2-fold elevation of high-density lipoprotein (HDL). These results suggested that CYP27A1 regulates cholesterol homeostasis, and alterations of its activities may subsequently lead to atherosclerosis[51].
ST6GALNAC5: The ST6GALNAC5 gene is the newest addition to the group of causal genes for familial CAD. InanlooRahatloo et al[52] studied a highly inbred Iranian pedigree of AD premature CAD. Unbiased GWAS combined with whole-exome sequencing of two affected members identified a polymorphism, G295A, in the ST6GALNAC5 gene that resulted in a p. Val99Met mutation. Targeted sequencing of all of the available members confirmed the co-segregation between this variant and the CAD phenotype. A search of ST6GALNAC5 mutations in other Iranians with confirmed CAD revealed a p.*337Qext*20 mutation in two unrelated patients with CAD (2 out of 160). Interestingly, one of the patients who carried this p.*337Qext*20 stop-loss mutation had one sibling with CAD and two unaffected siblings; a genetic analysis of the family again showed co-segregation of the mutation with disease status. ST6GALNAC5 encodes sialyltransferase 7e, a member of the sialyltransferase family. Sialyltransferases add sialic acids (acetylated derivatives of neuraminic acid) to the termini of carbohydrate chains in glycoproteins and glycolipids. Elevated sialyltransferase activity in blood cells and serum sialic acid levels[53] are associated with atherosclerosis and CAD. In vitro functional studies of proteins encoded by these two mutated ST6GALNAC5 genes revealed a two-fold increase in sialyltransferase 7e enzymatic activity[52]. Given that: (1) an un-biased approach was applied in the identification of the ST6GALNAC5 mutation in a large CAD pedigree; (2) convincing evidence demonstrates the co-segregation of the ST6GALNAC5 mutation with the familial CAD/MI phenotype; (3) additional functional mutations have been identified in the ST6GALNAC5 gene in unrelated CAD/MI patients and families; (4) no functional variations were identified in affected family members and unrelated controls; and (5) the available evidence supports the notion that sialic acid and sialyltransferase activity are involved in the pathogenesis of atherosclerotic arterial disease, it is reasonable to conclude that gain-of-function mutations in ST6GALNAC5, such as p. Val99Met and p.*337Qext*20, are monogenic causal genes for CAD. The prevalence of functional ST6GALNAC5 gene mutations in the general population and in patients with CAD is unknown. The mechanism by which mutant ST6GALNAC5 causes atherosclerosis and CAD and its potential role in targeted therapy or in the prevention of CAD remain unknown.
Serum lipid levels, particularly elevated LDL cholesterol and HDL, are important risk factors for the development of atherosclerotic CAD. Mutations in genes involved in lipid metabolism have been identified and demonstrated to be causal for dyslipidemia and related atherosclerotic CAD and MI with variable penetration. These genes and their mutations in association with CAD and MI have been extensively reviewed in the literature[54] and are presented in Table 2. Depending on the primary abnormality in lipid metabolism and its effects on CAD, monogenic lipid disorders can be categorized into primary elevated LDL (LDL receptor, ApoB-100, PCSK9, and LDLRAp or ARH), a primary reduction of HDL (ApoA1 in primary hypoalphalipoproteinemia, ABCA1 in Tangier disease, and the lecithin:cholesterol acyltransferase (LCAT) gene in Norum disease and Fish-Eye disease), and primary elevated triglycerides (TGs) (LPL, ApoC-II in type Ib hyperlipoproteinemia and the ABCG5/8 gene in Sitosterolemia).
Genes and variants that are the primary cause of high LDL cholesterol: Mutations in genes encoding the LDL receptor, apolipoprotein B-100 (an LDL receptor ligand) and the pro-protein convertase subtilisin kexin type 9 (PCSK9) cause AD familial hypercholesterolemia (FH). Patients who harbor homozygous [low-density lipoprotein receptor (LDLR)] mutations (1 in 1 million) display a 6- to 10-fold increase in plasma LDL-C from birth and experience CAD/MI in early childhood. The early atherosclerosis observed in children who are homozygous for FH is not associated with any other risk factors that suggest that elevated LDL alone can produce atherosclerosis in humans. Carriers of heterozygous LDLR mutations, demonstrating a frequency of 1/500 in the general population, display a 2-fold increase in low-density lipoprotein cholesterol (LDL-C) levels from birth and are at risk to suffer CAD and MI at 30s years of age. Approximately 5% patients with CAD and MI under the age of 60 years carry heterozygous LDLR mutations. A total of 1741 sequence variants (1122 unique variants) have been recorded in the British Heart Foundation LDLR database[55] (http://www.ucl.ac.uk/ldlr/Current/summary.php?select_db=LDLR&show=sum). These mutations are present in the form of exonic substitutions, small exonic rearrangements, large rearrangements, promoter variants, intronic variants, variant sin the 3’ untranslated sequence, point mutations, splice site mutations, and large deletions. These mutations are equally distributed throughout the gene[56,57]. Genetic testing of all known LDLR variants is available. This test is often considered as the first step in a stepwise genetic analysis for FH followed by tests to assess the ApoB-100 and PCSK9 genes[58].
Apo B-100 is a unique protein component in lipoproteins originating from the liver (VLDL, IDL, and LDL). Apo B-100 is also required for the synthesis, assembly, and secretion of hepatic TG-rich lipoproteins, and it binds to heparin and various proteoglycans found in arterial walls. The most important function of Apo B-100 is to bind the LDLR via its LDLR-binding domain to mediate the clearance of LDL from plasma. Two mutations in the Apo B-100 gene, C10580G (p. Arg3527Gln)[59] and C10800T (p. Arg3531Cys), result in the alteration of LDLR binding affinity. In addition, these mutations cause familial ligand-defective hypercholesterolemia (OMIM 144010) and are associated with early atherosclerotic arterial disease[60]. The frequency in the unselected general population of the Arg3527Gln and Arg3531Cys mutations is approximately 1 in 500 and 1 in 3000, respectively. Most recently, by taking advantage of whole-exome sequencing and linkage analysis of an AD hypercholesterolemia pedigree, a third mutation (p. Arg50Trp) was identified[61].
Linkage analysis of two large French ADH pedigrees resulted in the identification of two mutations in the PCSK9 gene (1p34.1-1p32), which encodes a protein that is also known as a neural apoptosis-regulated convertase 1 (NARC-1)[62]. A total of 9 gain-of-function mutations in PCSK9 genes in families with ADH have been reported[63]. These mutations cause a decreased number of LDLR, elevated levels of serum total and LDL cholesterol, and phenotypes of tendon xanthomas, premature CAD, MI and stroke. SNP rs11206510 (risk allele T) located in the PCSK9 gene is also associated with an increased risk of CAD and MI in an unbiased GWAS study[64]. However, loss-of-function mutations in PCSK9 identified by exome sequencing of individuals with extremely low LDL levels in the Atherosclerosis Risk in Communities study (ARIC) and Dallas Heart Study cohorts revealed that these mutations led to hypocholesterolemia and were protective against CAD and MI[65]. Secreted PCSK9 protein functions as an LDLR chaperone, binds to the EGF-A domain of the LDLR, decreases receptor recycling to the cell surface and promotes lysosomal degradation. Although the contribution of the PCSK9 gain-of-function mutation in ADH is rather small (< 3%), elucidation of the PCSK9 gain-of-function in ADH has shed light on the potential for the development of cholesterol-lowering agents by reducing the circulatory level of PCSK9 (PCSK9 inhibitors). These discoveries have resulted in the development of PCSK9 inhibitors as novel cholesterol-lowering agents.
Approximately 50 individuals of Mediterranean or Middle Eastern origin carry homozygous mutations in the autosomal recessive hypercholesterolemia (ARH) gene. ARH (1p34-1p35) was subsequently cloned and named LDL receptor adaptor protein 1 (LDLRAP1), which encodes a phosphotyrosine-binding domain protein and is required for LDLR internalization in hepatocytes. Two mutations, 432insA in exon 4 causing FS170Stop (ARH1) and the nonsense mutation G65A in exon 1 (p. trp22ter), account for most of the known cases of ARH in Sardinia[66]. The third mutant was a result of an ancient recombination between ARH1 and ARH2. In addition, 4 Italian ARH individuals from the mainland carried homozygous ARH1. Overall, ARH mutations are rare[67].
Genes and variants that are primary causes of low HDL cholesterol: Approximately 40% of patients with CAD have a low level of high-density lipoprotein cholesterol (HDL-C; < 40 mg/dL per current guidelines or age and sex-adjusted plasma HDL-C levels below the 10th percentile). Prospective cohort studies also suggest that low HDL-C is a significant, independent risk factor for CAD. An estimated 50% to 70% of the variations in HDL-C in the human populations are due to genetic factors, and the majority remain undefined[68].
Apolipoprotein A1 (Apo AI) is the major apolipoprotein in HDL-C and is a key determinant of the levels and metabolism of HDL-C. Apo AI functions as a cofactor for LCAT, which is responsible for the formation of most cholesterol esters in the plasma. Apo AI also promotes the efflux of cholesterol from cells. ApoAI mutations cause AD familial hypoalphalipoproteinemia. The homozygous loss of ApoAI leads to a complete absence of Apo AI and HDL-C levels < 5 mg/dL with normal LDL-C and TG levels. Heterozygous LoF Apo AI carriers have HDL-C levels that are approximately 50% less than normal HDL-C levels. ApoAI gene polymorphisms are associated with decreased HDL and an increased risk of premature CAD[69]. Yamakawa-Kobayashi et al[70] analyzed sequence variations in the ApoAI gene in Japanese children with low levels of HDL (below the first percentile in the general population) and found 3 frameshift and 1 splice site mutation with possible deleterious effects. They estimated the frequency of hypoalphalipoproteinemia due to ApoAI mutations to be 6% in subjects with low HDL cholesterol and 0.3% in the general Japanese population[70]. The A164S variant of the ApoAI gene identified by sequencing of the ApoAI gene in 190 Copenhagen City Heart Study participants predicts an increased risk of IHD [hazard ratio (HR) 3.2, 95%CI: 1.6-6.5], MI (5.5, 95%CI: 2.6-11.7) and overall mortality (2.5, 95% CI: 1.3-4.8). Despite comparable levels of plasma lipids and lipoprotein, including HDL-C and ApoAI, in A164S heterozygotes, heterozygous A164S carriers exhibit a decrease in survival by more than 10 years (P < 0.0001) compared with non-carrier controls[71,72]. In addition, two ApoAI variants (ApoAIParis and ApoAIMilano) were associated with a reduced risk of CAD, suggesting the occurrence of cardioprotective effects[73].
The ATP-binding cassette transporter (ABCA1) is involved in the initial phase of reverse cholesterol transport and the egress of free intracellular cholesterol and phospholipids from extrahepatic cells. Homozygous LoF ABCA1 variants are causal factors for the rare Tangier disease, which results in extremely low HDL-C levels, a 40% reduction of LDL-C compared with the general population, and an increased risk of early CAD[74,75]. A total of 200 LoF mutations in the ABCA1 gene have been reported (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=ABCA1, last accessed on August 6, 2014). Heterozygous carriers of the ABCA1 mutation exhibit an approximately 50% reduction in HDL-C without alterations in the levels of LDL-C and an increased risk of premature CAD[76]. The frequency of heterozygous carriers of ABCA1 mutations is estimated as approximately 3:1000 in the general population[77]. The R219K polymorphism in the ABCA1 gene is associated with a reduced risk of CAD, suggesting that this polymorphism provides protective effects against the disease[78].
LCAT catalyzes the esterification of free cholesterol with acyl groups derived from lecithin as an essential step in the maturation of HDL-C. Homozygous LoF in the LCAT gene causes rare autosomal recessive Norum disease with very low HDL (< 5th percentile), elevated TGs and decreased LDL-C. Greater than 80 genetic variants in the LCAT gene have been identified and reported to be associated with 29% of the individuals with low levels of HDL-C in the Netherlands[79]. However, the association between low levels of HDL-C caused by LCAT deficiency and an increased risk of CAD is not as certain as the associated risk of ApoAI[80]. This phenomenon may be explained by the observation that LCAT deficiency mainly causes decreased levels of ApoAII. HDL particles containing ApoAI, but not ApoAII, possess “anti-atherogenic” effects.
Genes and variants that are primary causes of elevated TGs: Plasma TGs predominantly occur in the form of intestinally synthesized chylomicrons (CMs), remnants in the postprandial state and hepatically synthesized VLDL in the fasted state. Plasma TG levels are a polygenic trait and is influenced by environmental factors and lifestyles, i.e., diet, physical activities and tobacco use. Large epidemiological studies have demonstrated that plasma TG concentrations are a strong independent risk factor for CAD[81].
Lipoprotein lipase (LPL) is the rate-limiting enzyme in converting VLDL to LDL. Homozygous LoF mutations in LDL genes cause LPL deficiency in rare (approximately 1 in a million) AR type I hyperlipoproteinemia characterized by marked hypertriglyceridemia with a decrease in HDL and LDL, eruptive xanthoma, hepatosplenomegaly, recurrent pancreatitis, and in some cases, premature atherosclerotic arterial disease. Greater than 100 LoF variants have been identified[82]. Sequencing data have suggested that rare LPL variants are actually common in patients with elevated TG levels[83]. Variants of LPL genes that are negatively associated with plasma levels of TG, positively associated with HDL and inversely associated with a risk of CAD have also reported, suggesting potential protective genetic variants against CAD[84]. ApoC-II is an activator of LPL. A homozygous LoF ApoC-II deficiency results in rare AD type Ib hyperlipoproteinemia with extremely elevated TG and chylomicron levels in the plasma, causing recurrent pancreatitis and, in some cases, ApoCIISt. Michael (Gln70Pro)[85] and other conditions[86], leading to premature ischemic vascular disease. The significance of these mutations in general population remains to be explored.
Sitosterolemia is characterized by hyperabsorption and the retention of dietary cholesterol and sterols, including plant and shellfish sterols, leading to high levels of plant sterols in the plasma, the development of tendon and tuberous xanthomas, accelerated atherosclerosis, and premature CAD. LoF mutations in ABCG5 (encoding sterolin-1) and ABCG8 (encoding sterolin-2) cause sitosterolemia. All of the probands identified in the sitosterolemia pedigree have homozygous mutations in either ABCG5 or ABCG8[87]. The prevalence of ABCG5 and ABCG8 heterozygous carriers and their effects on cholesterol metabolism and atherosclerotic disease in the general population remain unclear.
Monogenic traits and their causal genetics only explain a small proportion of the genetics of CAD and MI. Prior to the completion of human genome sequencing, CAD and MI gene discovery largely employed pedigree-based linkage analysis and positional cloning with the limited availability of genomic markers. The Human Genome Project, HapMap project, and 1000 Genomes Project provided a reference of 3.2 billion nucleotide base pairs of the human genome and 3 million single nucleotide polymorphisms (SNPs) distributed throughout the genome. These SNPs serve as high-density genomic markers for the entire genome. The developments of high-density microchips containing millions of SNPs, high-throughput analytic technology, and powerful biostatistics data mining tools have permitted genome-wide association studies (GWAS). GWAS is a non-hypothesis-driven, unbiased analysis of the potential associations between traits of interest (disease, phenotypes, etc.) and genomic markers (SNPs) consisting of tens of thousands of cases and controls[88]. In 2007, SNPs located in 9p21 were identified as strongly associated with CAD and MI based on the results of four nearly simultaneous publications. Numerous GWAS studies have been subsequently conducted, involving tens of thousands of CAD and MI cases and controls inclusive of a large spectrum of demographic, geographic and ethnic backgrounds. The largest meta-analysis of GWAS data reported by the CARDIoGRAMplusC4D Consortium included a total of 63746 CAD cases and 130681 control subjects and confirmed/identified 46 CAD susceptibility loci[84]. Together with a 6q21 locus that was identified in the Chinese Han population by Wang et al[89] and an additional 3 CAD susceptibility variants identified by IBC 50K[90], which were not confirmed in the CARDIoGRAMplusC4D Consortium study, a total of 50 GWAS-identified CAD susceptibility genomic loci were reported. Recent reviews have focused on GWAS discovery of CAD and MI susceptibility loci[91-95].
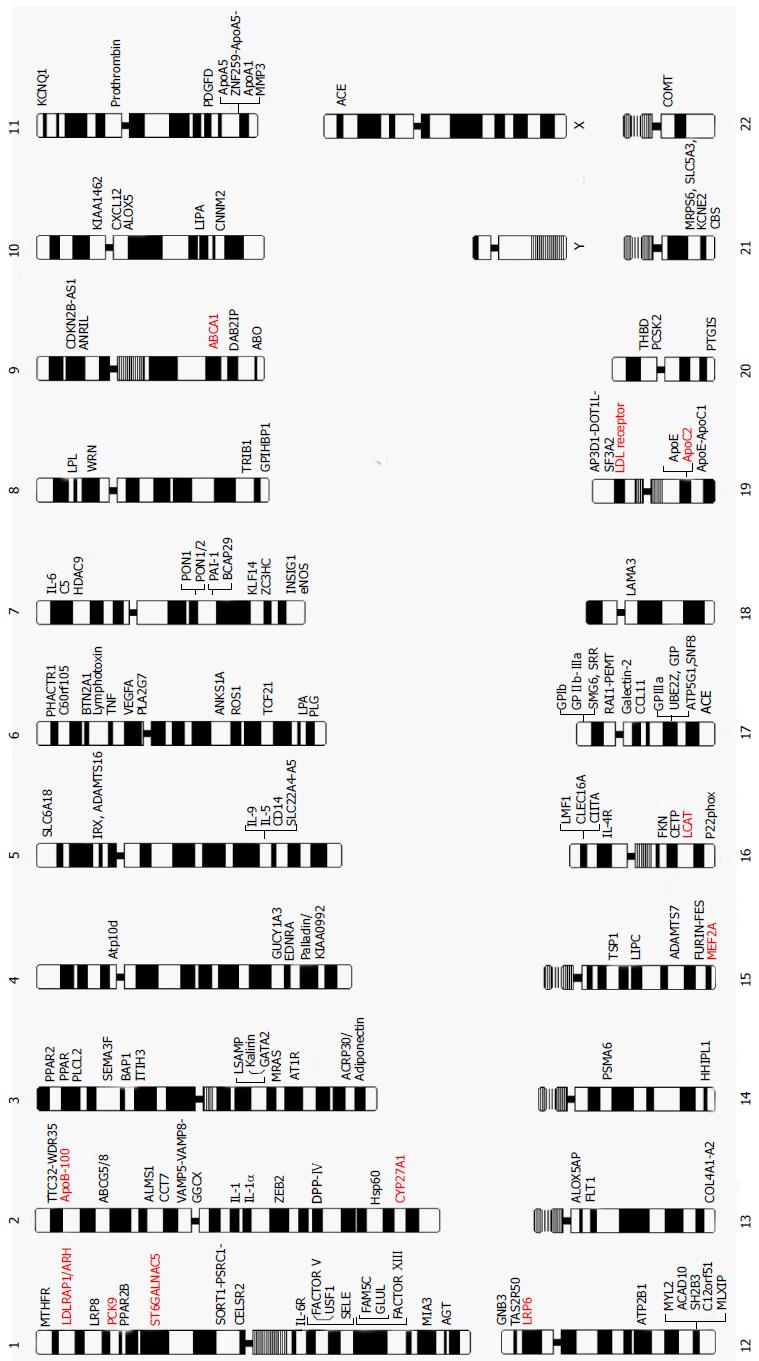
The journey to understand genetic/genomic architecture of CAD and MI continues with steady progress. Figure 2 provides a complete map of the genes and loci that are reported in literature to be causal for, susceptible to, or associated with the risk of CAD and MI identified by either linkage analysis and/or association analyses with either candidate-gene or genome-wide approaches. The genes and mutations that are potentially causal for monogenic CAD and MI families are also presented in Table 2.
The ultimate goals of the study of the genetics of CAD and MI are to reveal genes and their products involved in the development of CAD and MI, understand the molecular and cellular pathophysiology and subsequently establish risk stratification strategies to direct prevention and also develop effective therapeutic approaches. Despite the total of 147 genes (Figure 2) with variants that are causative for or associated with CAD and MI, these genes only explain less than 20% of the heritability of CAD and MI. Furthermore, the biological functions and pathophysiological roles of most of the gene variants and genomic loci are not fully understood. Early attempts to use genetic risk markers of CAD to predict long-term outcomes were not successful, and many challenges remain before the ultimate goal of understanding the genetics of CAD can be realized.
Searching for unexplained heritability: GWAS is based on the hypothesis of a “common disease, common variant”. The sensitivity of GWAS in detecting a significant association between genetic variants and traits is limited to high frequency variants (5%)[96]. Regarding CAD and MI, most of the GWAS-identified variants individually or in combination confer relatively small increments in risk (1.1- to 1.5-fold) and explain only a small proportion of heritability. The well-recognized sources for the missing heritability of complex traits[97-99] include: (1) larger numbers of variants with a smaller effect that remains to be identified; (2) a low minor allele frequency (0.5%-5%) or rare variants (< 0.5%) with a larger frequency that are not detectable using the available arrays; (3) structural variants (i.e., copy number of variants due to insertion or deletions, inversions, or translocation) are poorly captured by SNPs; (4) a low power to detect gene-gene interactions; and (5) an inability to detect gene-environment interactions using the current GWAS methodology. Strategies have been suggested and explored to overcome these pitfalls, including: (1) analyzing phenotypically well-defined cases and controls[100], increasing the numbers of participants[84], and utilizing extreme phenotype groups[54,101]; (2) developing powerful biostatistics tools to enrich the signal and detect sensitivities and to capture the additive effects of variants, gene-gene interactions and rare variants[102,103]; (3) integrating available functional information, i.e., eQTLs, protein structure/function predictions, and known pathways and networks related to the traits, to prioritize GWAS signals[104] and performing integrative analyses[105]; (4) customizing fine mapping SNPs or use next-generation sequencing regions of interest to capture rare variants and structural variants; and (5) considering the epigenomic regulation of gene expression.
Many current phenotypes are subjectively measured and may represent numerous underlying biological processes. Misclassifying a phenotype can reduce power in GWAS relative to expectations based on power calculations of idealized homogeneous populations. Strong genotypic effects that are important in a small homogeneous subgroup could have a small or even negligible effect within an entire population[106].
Functional annotation of known genes and variants: Numerous genes and variants associated with CAD and MI have been explored in the last two decades. Candidate gene approaches (positional cloning, linkage analysis, and candidate-gene association analysis) provide a direct link between the pathogenesis of CAD/MI and candidate genes. Variants or chromosomal loci identified by GWAS however do not associate with specific genes or pathways. Only one-third of the 45 CAD loci reported in the largest CAD and MI GWAS study contain a known functionally relevant candidate gene[84]. The 9p21 locus is the CAD locus discovered by GWAS and remains the strongest association with CAD in the human genome. However, the SNPs defining the 9p21 association with CAD are all located in intergenic locations rather than in coding or regulatory regions. Functional annotation of the 9p21 locus in association with CAD has focused on the two closest protein-coding genes, CDKN2A and CDKN2B, and an additional CDKN2B antisense noncoding RNA (ANRIL). The systematic functional annotation of these CAD and MI gene variants and loci will provide insight regarding the pathophysiology of the disease. These functional studies will require a combination of tissue, cell and animal model systems as well as assessments at the levels of gene expression, protein modification and metabolism[107,108].
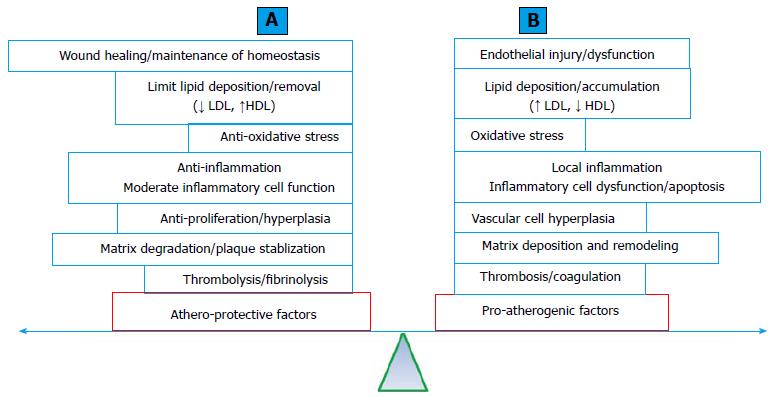
Protective genetic factors against CAD and MI: Atherosclerosis is a multi-decade pathological process. A simplified pathologic process (Figure 3A) includes: (1) endothelial injury; (2) lipid particle deposition (fatty streak formation); (3) local cellular and inflammatory responses (early atheroma formation). The process is followed by (4) atheroma progression with the formation and expansion of the necrotic core, fibrous cap, matrix accumulation and various degrees of plaque instability; and (5) intertwined with various degrees of thrombosis formation[109]. The standard human genetic approach to CAD involves searching for genetic risk factors or susceptibility genes for the disease. Could genetic factors be protective against atherosclerosis and CAD and MI? Protective genetic factor against CAD and MI may antagonize the effects of genetic risk factors and potentially explain a portion of the missing heritability. The presence of protective genetic factors will alter the predictive value for CAD and MI using risk variants. The identification of protective genetic factors may shed light on the mechanisms of CAD and MI and further guide the development of strategies or therapy.
Clinical practice often encounters individuals of an advanced age with multiple TRF for CAD and MI without demonstrated angiographic evidence of CAD. The offspring of centenarians have a significantly reduced rate of cardiovascular complications[110]. The current understanding of the pathophysiology of atherosclerosis and CAD and MI supports the notion that the development and progression of atherosclerosis is an accumulating effect of the imbalance between athero-protective and pro-atherogenic processes (Figure 3B). Approximately 500 genes reported in the literature have been tested for their effects on atherogenic processes in atherosclerosis-prone mouse models (ApoE deficiency or LDL receptor deficiency with a high-fat diet) with transgenic (gain-of-function), knockout (LoF) or both genetic modifications. LoF mutations in approximately half of these genes accelerate atherosclerosis in a mouse model, whereas gain-of-function mutations significantly reduced atherosclerosis. It is plausible to suggest that these genes normally function as protective factors against atherosclerosis. In contrast, the remaining half of these reported genes exert pro-atherogenic effects that are normal or consistent with their gain-of-function mutations[111] (a complete list of these genes is available from the authors upon request).
Most genomic association studies have been designed to identify CAD/MI susceptibility genes or polymorphisms. The largest genetic study to assess the impact of common genomic variation on the risk of CAD reported a total of 45 CAD susceptibility loci and an additional 104 likely independent SNPs that were associated with an increased risk of CAD and MI, explaining approximately 10.6% of the heritability. No protective variants were reported[84]. Candidate gene-based association studies identified polymorphisms that are significantly associated with a reduced risk of CAD and MI (Table 3). However, the results obtained for many of these potentially protective genetic loci against CAD are conflicting.
| Chr | Gene | Protective alleles | Ref. |
| 1p13 | Rs599839 A>GRs646776 T>C | C/G haplotype | [161,162] |
| 1q22 | E-selectin | G2692A; C901T | [163] |
| 1q31 | GLUL | Rs10911021 T>C, TT allele | [164] |
| 1q31 | IL-10 | G(-1082)A, GG genotype | [165] |
| 1p34 | LRP 8 | TCCGC | [166] |
| 2p21 | ABCG 5/8 | Rs41360247 | [167] |
| 3p25 | PPARγ2 | Pro12Ala homo | [168-170] |
| 3p25 | PPARγ | C161T | [171] |
| 3q27 | Adiponectin | Rs1501299 (G276T), TT allele | [172-174] |
| 8q21 | FABP4 | Rs77878271 | [175] |
| 6p12.3 | PLA2G7 | R92H | [176] |
| 6p25.3 | FXIII | Val34Liu | [177,178] |
| 7q21.3 | PON1/2 | Gln192Arg | [112,116] |
| 7q32.3 | KLF14 | Rs4731702 T/T allele | [179] |
| 7q36 | INSIG1 | Hap3 (T/G/A) | [180] |
| 9q31.1 | ABCG1 | G1051A, r219K, KK allele | [78] |
| 11q23.3 | APOC3 | R19X | [181] |
| 13q34 | FVII | R353Q; QQ allele A2 allele (without a 10 bp insertion) | [182] |
| 16q13 | FKN | T280M allele; Rs4329913; Rs7202364 | [183,184] |
| 16q24 | NADPH p22phox | C242T | [185,186] |
| 17p13.2 | GP1bα | Thr/Th; TT haplotype | [187] |
| 21q22.1 | MRPS6 | C699T (TT) or T1080C (CC) | [64] |
HDL provides protection against atherosclerotic CAD partially through its anti-oxidative effects. Serum paraoxonase is responsible for most of the antioxidant properties of HDL. Human paraoxonase is encoded by the family of PON1, PON2 and PON3 genes. Low serum PON1 activity is associated with an increased risk of CAD and its severity[112,113]. Many candidate gene association studies have revealed that PON1 (Leu55Met, Gln193Arg) and PON2 (Ser311Cys) polymorphisms are associated with the risk of CAD[114] and its angiographic severity[115]. For example, an association study in a single center of consecutive patients who underwent coronary angiography revealed a significant dose-dependent association of the PON1 genotypes (192 Q/R) and serum PON1 (QQ192 > QR192 > RR192) as well as an inverse association with systemic indices of oxidative stress. In addition, 192 Q (QQ and QR) was associated with a decreased risk of cardiovascular and all-cause mortality[116]. The PON1/PON2 haplotype comprising M55, Q1192 in PON1 and Cys 311 in PON2 is associated with a significant protective effect against the risk of MI[117]. PON1-deficient mice in an ApoE-/- background fed a high-fat diet exhibit significantly exaggerated atherosclerosis compared with ApoE-/- mice carrying the wild type PON1 gene[118,119]. Germline transgenic or transient adenoviral vector-mediated overexpression of atheroprotective PON1 (55L/192Q) in ApoE-/- mice revealed protective effects against atherosclerosis with ApoE-/- without transgenic PON1[120,121]. Multiple layers of evidence suggest that genetic polymorphisms in PON1 and PON2 lead to an increase in serum paraoxonase activity that may provide protective effects against CAD. However, the frequency of these variants in the general population remains to be determined.
Primary prevention of CAD is gauged based on the risk categories derived from the risk assessment with TRFs, such as the Framingham risk score (FRS) in the United States[122], the SCORE risk equation in Europe[123], the Reynolds risk score for women[124] and the PROCAM risk score in Germany[29]. The discovery of causal genetic factors for monogenic CAD and MI, such as monogenic lipid disorders, have made it possible to perform clinical genetic screening of family members and to provide enhanced primary prevention to carriers of causal mutations. This approach has been shown to be cost-effective[125]. The association between genetic polymorphisms and the risk of CAD and MI provides an opportunity to use genetic information and develop a GRS to improve the risk prediction of CAD and MI in the general population and subsequently guide preventive strategies. The GRS is calculated either in an un-weighted manner by adding allele numbers (0 for no risk allele, 1 for one allele and 2 for both alleles) with a weighted GRS typically by using the reported effect sizes from the reference studies as weights for the risk allele counts or with a weighted GRS mean, which is derived by dividing the sum of the weighted GRS allele counts by the number of the SNPs. The association of GRS with the risk of the CAD endpoint has been assessed. Thus, GRS is evaluated if the addition of GRS to the traditional risk scoring model improves the discrimination measured using AUC or C statistics or results in risk category net reclassification improvement (NRI). To be clinically applicable, GRS must eventually be validated in independent prospective studies. The expectations are high[126]. Research on this topic is growing. The scientific community has provided guidelines regarding the design, performance and reporting of studies investigating genetic risk prediction[127,128]. However, the outcomes have been mixed.
Early in 2004, Humphries et al[126] reported a non-significant improvement of the risk prediction power to PROCAM risk score with the addition of ApoE genotype information in the Northwick Park Heart Study II (NPHSII) cohort (ROC increased from 0.65 to 0.67, P = 0.11). The addition of genetic variants of IL6 and PPARα did not result in any improvement of the CAD prediction power of the PROCAM score[126]. Chromosome 9p21.3 has demonstrated the strongest association with CAD in GWAS studies. The addition of the genotype of SNP rs10757274 A>G in the 9p21.3 locus did not significantly improve the predictive value of the FRS, but it improved the reclassification of coronary heart disease (CHD) risk and guided primary prevention for a high-risk population in a prospective study[129]. This conclusion was subsequently confirmed in studies using independent cohorts[130-133]. Statistical modeling revealed that larger numbers of genetic variants, higher odds ratios (OR) and the genotype frequency of individual variants can improve the discriminative accuracy of area under the receiver operating characteristic curve (AUC) using the genetic score to predict the risk of CAD and MI[134,135]. The application of 100 established variants with ORs ranging from 1.13 to 1.42 can achieve an AUC of 0.76, which is comparable to most of the currently used conventional risk scoring systems[135]. A rapid increase in the number of studies reporting the development and validation of GRS to predict the risk of CAD has been recently noted[136,137].
Morrison et al[138] calculated the GRS based on the number of risk alleles of 11 CAD-associated SNPs identified in the Atherosclerosis Risk in Communities Study (ARIC) cohort and combined the results with the ARIC Cardiovascular Risk Score (ACRS) to predict CAD. These researchers found that the addition of GRS to the traditional risk score significantly increased the AUC to predict the risk of CAD in blacks and suggested improved CAD risk prediction in whites[138]. In a large prospective cohort study with a median of 10.7 years of follow-up, Ripatti et al[139] found that individuals with a GRS in the top quintile derived from 13 multi-locus SNPs of CHD exhibited a 1.66-fold increased risk of CHD adjusting for TRF. However, the GRS did not improve the C index over the TRFs and family history or the net reclassification of risk categories. Paynter et al[140] prospectively studied GRS from 101 SNPs in the large Women’s Genome Health Study with a median follow-up of 12.3 years and found that the GRS did not improve the discrimination or reclassification of the ATP III risk score. Most recently, by choosing SNPs that were repeatedly and reproducibly confirmed in multiple GWAS studies using improved statistics tools and systematic risk stratifications of TRFs, GRS added significant predictive value to improve risk predictions[141-146]. For example, Thanassoulis et al[142] constructed a GRS with 13 CAD risk SNPs and assessed participants in the FHS. These researchers not only confirmed the association between GRS and incident CHD and a high coronary artery calcium score (CAC) but also demonstrated that GRS modestly but significantly improved the risk reclassification for incident CHD and significantly improved the discrimination for a high CAC[142]. However, the addition of 16 newly discovered SNPs to the GRS (total of 29 SNPS) did not improve the performance of the GRS in contrast to previous in silico computations[135]. Tikkanen et al[146] derived a weight GRS using 28 SNPs associated with risk for CAD and MI in the large FINRISK study cohort with up to 19 years of follow-up for CHD. These researchers discovered a highly significant independent association between GRS and the risk of CHD. The addition of GRS to TRF with and without a family history significantly improved both the risk discrimination for all end points and the reclassification of individuals in the intermediate-risk category (clinical NRI = 27%). Similar results were validated in additional independent cohorts. Furthermore, this GRS was used as a novel risk marker in the 2-stage population screening study Emerging Risk Factors Collaboration. The addition of GRS screening of individuals with intermediate risk per TRF screening reclassified 19% of the group into the low- and 12% into the high-risk category, who thus became eligible for more aggressive primary prevention[146].
GRS derived from genetic variants associated with TRF for CAD is generally confirmed by an association with the disease, but it does not improve the discrimination of CAD and MI derived from the TRF assessment[142]. Kathiresan et al[147] calculated genetic scores for 5414 subjects in the Malmo Diet and Cancer Study based on the number of unfavorable alleles of nine SNPs with associations with LDL or HDL cholesterol levels. In this study, the genetic score was an independent risk factor for incident CVD over a median of 10.6 years of follow-up and modestly improved the clinical risk reclassification (Adult Treatment Panel III, ATP III classification) for individuals in the intermediate-risk category (26% rate of reclassification). However, this genetic score did not improve the risk discrimination[147]. Isaacs et al[148] derived a GRS from 95 blood lipid loci with common genetic variants with confirmed cumulative effects on subclinical atherosclerosis and clinical CAD and MI, but the score did not improve the clinical AUCs in combination with FRS. Similarly, Guella et al[149] analyzed a weighted GRS based on the top SNPs in 12 loci in the hemostatic gene pathway and found a 2.69-fold increased risk of early onset MI in subjects in the highest GRS quintile compared with those in the lowest quintile[149]. The predictive value of this weighted GRS has not been studied in a prospective study.
Compared with traditional risk assessment, the advantages of GRS are evident and include the following characteristics: (1) GSR is highly stable over a life time. This information permits the early identification of individuals who are at risk and the implementation of early intervention; (2) Current technology allows the simultaneous measurement of large numbers of genetic variants; (3) The presence of specific genetic risk alleles may provide information regarding targeted preventive intervention; and (4) Most of the SNPs identified by GWAS do not correlate with known TRFs. SNP-based GRS offers complementary information for risk prediction. GRS derived from CAD-associated SNPs provides significant additional predictive power that exceeds TRFs based on both AUC and NRI criteria. Genomic technology has also reduced the cost associated with genotyping a large number of SNPs. It is reasonable to predict that the GRS of CAD will eventually be a component of clinical practice. A number of questions remain to be addressed: (1) The potential difference of predictive values among candidate gene approach identified variants vs GWAS variants. Genetic association studies using a candidate gene approach often consist of a small sample size and cannot be replicated in different populations. The minimal criteria for a genetic variant to be included in CVD clinical risk management is recommended, including a meta-analysis based on the data from a minimum of three different independent studies that comprise at least a total of 1000 cases[126]. Potential causal variants for familial CAD and MI (Table 2) are low frequency, high impact variants. The allele frequencies in the general population remain to be determined. The appropriate techniques to incorporate these variants into the GRS remain to be addressed; (2) Protective genetic variants against CAD and MI can potentially attenuate predisposing effects of risk alleles. The number and frequency of protective genetic factors against CAD and MI in the population remain be determined. It will be interesting to evaluate how these protective variants influence the GRS calculation and its predictive power; (3) Gene-gene interactions: The synergistic effects between genetic variants have been reported in association with the risk of CAD[150,151]. Consideration of the combined effects in the GRS model may facilitate risk prediction; and (4) Gene-environmental interactions. The effects on the risk of CAD and MI by certain environmental factors depend on genetics in a “context dependency” fashion. In addition to the overall calculated GRS, information about specific genetic variants may guide personalized preventive intervention. For example, the information obtained for the ApoEe4 allele is associated with exaggerated CAD and MI risk in tobacco smoker but not in non-smoker. It would be particularly important to advise smoking cessation in ApoEe4 carriers[126]. Although, the inclusion of ApoEe4 in the GRS calculation may overestimate the risk for non-smokers.
Percutaneous coronary intervention (PCI), an effective and safe alternative treatment modality for obstructive CAD, has become one of the most commonly performed therapeutic medical procedures since it was first performed by Grüntzig et al[152] in 1977 (http://www.ptca.org/nv/timeline.html. Last accessed on 8/30/3014). Restenosis, which is defined as a renarrowing of the treated vessel area that equals or exceeds 50% of the lumen in the adjacent normal segment, is an entity that is produced with the birth of PCI. The process often results in recurrent symptoms that require repeated intervention. Early experiences in balloon angioplasty revealed a restenosis rate of greater than 50%. The implantation of bare metal stents reduces the restenosis rate to 20% to 30%, mainly via the elimination of early elastic recoil and negative remodeling. The development of drug-eluting stents (i.e., silorimus and paclitaxel as prototype-eluting drugs) further reduced the rate to 5% to 15%, as demonstrated in large randomized controlled trials[153]. Despite the advancement of PCI equipment and technology, late luminal loss due to in-stent restenosis (ISR) remains the “Achilles heel” for interventional cardiologists treating CAD.
ISR is a complex disease. Patient factors, such as older age, hypertension, diabetes mellitus and a history of restenosis, increase the risk of ISR[154], whereas tobacco use decreases the risk. Lesion characteristics, such as chronic total occlusion, a small vessel diameter, long lesions, the degree of calcification, ostial/bifurcation lesions, and restenosis lesions, are associated with an increased risk of restenosis. Procedural-related factors, such as multiple stents, bare metal stents, small diameter and/or long stents, stent fracture, under-expansion, and the presence of edge dissection, increase the risk of restenosis[155]. The pathophysiology of ISR is not fully understood. Compared with balloon angioplasty, stent placement achieves greater acute gain (greater lumen caliber), prevents acute elastic recoil and plaque prolapse, and reduces negative remodeling. However, greater injury to the deeper arterial layers, de-endotheliarization and accumulating layer of platelets and fibrin on the stent surface in association with stent deployment trigger an increased inflammatory response and wound healing process, which includes leukocyte infiltration, cytokine and growth factor release, VSMC activation and proliferation, and matrix production. These processes result in neointimal hyperplasia, the main process of ISR, and neo-atheroma formation during late luminal loss[156]. It is well known that these processes, in particular the cell cycle regulation and inflammatory responses, involve a sophisticated regulatory network consisting of a multitude of proteins. The abundance, modification, and temporal and spatial expression of these proteins are controlled by genetic elements. Genetic factors are hypothesized to influence the risk of ISR for individuals undergoing PCI with stent placement.
Traditional epidemiological genetics has not yet established the heritability of ISR. Candidate gene association analyses using genetic markers, genomic polymorphisms or SNPs have revealed many important associations between genomic variants and the risk of ISR. Table 4 summarizes the published gene polymorphisms associated with the risk of ISR. These genes participate in the regulation of the cell cycle [CCNB1, p27kip1, eNOS, miRNA-146a and p53); inflammation (IL1B, IL1RN, IL8, IL18, TNFα, CD18, CD14, ICAM1 and CX37]; oxidative stress (RAGE, eNOS and HO-1); metabolism/hormonal regulation (ALOX5AP, vitamin D receptor (VDR), α-estrogen receptor (αER), methylenetetrahydrofolate reductase (MTHFR), adiponectin, UPC3, and FBG) and coagulation/thrombosis [factor V leiden, fibrinogen beta chain (FGB), GPx-1, PAI-1, and P2RY12]; epigenetic regulation of gene expression (KAT2B), matrix deposition and degradation (MMP12); and the renin-angiotensin-aldosterone system (RAAS) system (ACE and AGTR1), which is also related to the maintenance of vascular hemostasis (ICAM-1). Although most of the polymorphisms identified in these genes are associated with an increased risk of ISR, some genetic variants were found to have protective effects against ISR, such as the TT genotype in exon 11 of CD18 (ITGB2), the A allele of ALOX5AP, the A allele of the p27kip1 gene, the AA genotype at position -374 of the RAGE promoter, the CC alleles in miRNA-146a, and allele 2 (C allele) in the IL-1RN gene. There are discrepancies in the literature reporting candidate gene-based association analyses and unbiased GWAS studies that involve significantly heterogeneous cases and controls as well as relatively small sample sizes[157]. Further validation and physiological annotation of most of these associations between polymorphisms and the risk of ISR in future studies will be essential.
| Chr locations | Gene symbols | Genetic polymorphisms | Effects on risk of ISR | Pathway involved | Ref. |
| 1p36.3 | MTHFR | C677T | ↑ ISR | Metabolism | [188] |
| 1q32.1 | IL10 | G(-2849)A; G(-1082)A; A4259G | ↑ ISR | Inflammation | [189] |
| 1p35.1 | CX37 | C1019T | ↑ ISR in men | Inflammation | [190,191] |
| 2q14 | IL 1B | C(-511)T | ↑ ISR | Inflammation | [192] |
| 2q14.2 | IL-1RN | T8006C | ↓ ISR | Inflammation | [154] |
| 3p21.3 | GPx-1 | C599T (rs1050450) | ↑ ISR | Thrombosis | [193] |
| rs8179164 A>T | ↑ ISR | [194] | |||
| 3p24 | KAT2B | rs6776870 G>C; rs2929404 T>C; rs17796904 T>C; rs4858767 G>C | ↑ ISR | Epigenetic/gene expression | [194] |
| 3q24 | AGTR1 | rs5182 T>C | ↑ ISR | Vascular homeostasis | [194] |
| 3q24 | P2RY12 | P2Y12 Haplotype H1 (5 P2Y12 ht-SNPs) | ↑ ISR | Thrombosis | [195] |
| 3q27 | Adiponectin | T(+45)G rs2242766 | ↑ ISR | Inflammation | [196] |
| 4q13 | IL-8 | A(-251)T + C(781)T | ↑ ISRS | Inflammation | [197] |
| 4q28 | FGB | rs1044291 T>C | ↑ ISR | Thrombosis | [194] |
| 5q12 | CCNB1 | rs350099 C>T (TT); rs350104 T>C (CC); rs164390 T>G (GG); TT/CC/GG haplotype | ↑ ISR | Cell cycling | [198] |
| ↑↑ ISR | |||||
| 5q31.1 | CD14 | C(-260)T | ↑ ISR | Inflammation | [199] |
| 5q34 | miRNA-146a | rs2910164>G (G/C) | ↑ ISR | Inflammation | [200] |
| rs2910164>G (C/C) | ↓ ISR | ||||
| 6p21.3 | TNFα | T (-857)C +C(-1031)T | ↑ ISR | Inflammation | [201] |
| 6p21.3 | RAGE | T(-374)A | ↓ ISR | Inflammation | [202] |
| 6q25.1 | αER | PvuII (C/T) > (TT) | ↑ ISR in women | Cell cycling | [203] |
| 7q22.1 | PAI-1 | 5G/5G | ↑ ISR (smoker) | Thrombosis | [204] |
| ↓ ISR (nonsmoker) | |||||
| 7q36.1 | eNOS | 298C/T (p. Glu298Asp)(rs1799983>T); T(-786)C | ↑ ISR | Cell proliferation | [193,205,206] |
| 11q22.3 | MMP12 | rs12808148 C>T; rs17099726 G>T | ↑ ISR | Matrix deposition | [194] |
| 11q22.2 | IL-18 | G(-137)T | ↑ ISR | Inflammation | [207] |
| 11q13.4 | UPC3 | C(-55C)T | ↑ ISR | Metabolism | [208] |
| 12p13.1 | p27kip1 | (-838)AA | ↓ ISR | Cell cycling | [209] |
| 12q13.11 | VDR | Block 2 AA haplotype | ↑ ISR | Metabolism | [210] |
| rs11574027 T>G; rs11574077 G>A | ↑ ISR | [194] | |||
| 13q12 | ALOX5AP | rs10507391 T>A; rs17216473 G>A | ↑ ISR | Lipid metabolism | [211] |
| rs17222814G>A | ↓ ISR | ||||
| 17p13.1 | p53 | Arg72Pro | ↑ ISR | Cell cycling | [212] |
| 17q23.3 | ACE | D allele: no 287-bp Alu repeats insertion in intron 16 | ↑ ISR | Cell cycling | [29-32,213-215] |
| 19p13.2 | ICAM-1 | K469E | ↑ ISR | Cell-cell interaction | [216] |
| 21q22.3 | CD18 | C1323T | ↓ ISR | Inflammation | [217] |
| 22q13.1 | HO-1 | > 29 TG repeats in promoter | ↑ ISR | Oxidative stress | [218] |
Discoveries of genetic factors associated with the risk of ISR will not only provide insight regarding the molecular mechanisms underlying the pathogenesis of ISR but also facilitate the development of novel strategies or agents to prevent ISR. More importantly, a complete understanding of genetic risks for ISR will provide clinicians with prognostic information to tailor revascularization strategies, PCI with stenting or coronary artery bypass grafting (CABG). Understandably, patients, in particular younger patients with significant CAD who possess genetic risk factors for ISR, will theoretically benefit more from CABG, and patients carrying protective genetic factors for ISR may benefit from PCI stenting to avoid surgery.
P- Reviewer: Coccheri S, Rallidis LS S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28-e292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3046] [Cited by in F6Publishing: 3492] [Article Influence: 349.2] [Reference Citation Analysis (0)] |
| 2. | Lusis AJ, Fogelman AM, Fonarow GC. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 2004;110:1868-1873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 3. | Stokes J, Dawber TR. The silent coronary: the frequency and clinical characteristics of unrecognized myocardial infarction in the Framingham study. Ann Intern Med. 1959;50:1359-1369. [PubMed] [Cited in This Article: ] |
| 4. | Paton BC. The accuracy of diagnosis of myocardial infarction; a clinicopathologic study. Am J Med. 1957;23:761-768. [PubMed] [Cited in This Article: ] |
| 5. | Cybulsky MI. Morphing the topography of atherosclerosis: an unexpected role for PECAM-1. Arterioscler Thromb Vasc Biol. 2008;28:1887-1889. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Luo AK, Jefferson BK, Garcia MJ, Ginsburg GS, Topol EJ. Challenges in the phenotypic characterisation of patients in genetic studies of coronary artery disease. J Med Genet. 2007;44:161-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Thomas CB, Cohen BH. The familial occurrence of hypertension and coronary artery disease, with observations concerning obesity and diabetes. Ann Intern Med. 1955;42:90-127. [PubMed] [Cited in This Article: ] |
| 8. | Rose G. Familial patterns in ischaemic heart disease. Br J Prev Soc Med. 1964;18:75-80. [PubMed] [Cited in This Article: ] |
| 9. | Slack J, Evans KA. The increased risk of death from ischaemic heart disease in first degree relatives of 121 men and 96 women with ischaemic heart disease. J Med Genet. 1966;3:239-257. [PubMed] [Cited in This Article: ] |
| 10. | Schildkraut JM, Myers RH, Cupples LA, Kiely DK, Kannel WB. Coronary risk associated with age and sex of parental heart disease in the Framingham Study. Am J Cardiol. 1989;64:555-559. [PubMed] [Cited in This Article: ] |
| 11. | Nora JJ, Lortscher RH, Spangler RD, Nora AH, Kimberling WJ. Genetic--epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503-508. [PubMed] [Cited in This Article: ] |
| 12. | Anderson AJ, Loeffler RF, Barboriak JJ, Rimm AA. Occlusive coronary artery disease and parental history of myocardial infarction. Prev Med. 1979;8:419-428. [PubMed] [Cited in This Article: ] |
| 13. | Hamby RI. Hereditary aspects of coronary artery disease. Am Heart J. 1981;101:639-649. [PubMed] [Cited in This Article: ] |
| 14. | Chesebro JH, Fuster V, Elveback LR, Frye RL. Strong family history and cigarette smoking as risk factors of coronary artery disease in young adults. Br Heart J. 1982;47:78-83. [PubMed] [Cited in This Article: ] |
| 15. | Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A. Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol. 1984;4:793-801. [PubMed] [Cited in This Article: ] |
| 16. | Benedict RB. Coronary heart disease in identical female twins. Am J Med. 1958;24:815-819. [PubMed] [Cited in This Article: ] |
| 17. | Giknis FL, Holt DE, Whiteman HW, Singh MD, Benchimol A, Dimond EG. Myocardial infarction in twenty-year-old identical twins. Am J Cardiol. 1965;16:122-126. [PubMed] [Cited in This Article: ] |
| 18. | Sidd JJ, Sasahara AA, Littmann D. Coronary-artery disease in identical twins. A family study. N Engl J Med. 1966;274:55-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 19. | Kreulen TH, Cohn PF, Gorlin R. Premature coronary artery disease in identical male twins studied by selective coronary arteriography. Cathet Cardiovasc Diagn. 1975;1:91-96. [PubMed] [Cited in This Article: ] |
| 20. | Segura L, Moreno R, Macaya C. [Coronary artery disease and percutaneous coronary intervention in a set of twins]. Rev Esp Cardiol. 2007;60:86-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 21. | Cederlöf R, Friberg L, Jonsson E. Hereditary factors and “angina pectoris”. A study on 5,877 twin-pairs with the aid of mailed questionnaires. Arch Environ Health. 1967;14:397-400. [PubMed] [Cited in This Article: ] |
| 22. | Zdravkovic S. Coronary heart disease in Swedish twins: quantitative genetic studies. Thesis. Solna, Sweden: Karolinska Institutet 2006; . [Cited in This Article: ] |
| 23. | Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247-254. [PubMed] [Cited in This Article: ] |
| 24. | Wienke A, Holm NV, Skytthe A, Yashin AI. The heritability of mortality due to heart diseases: a correlated frailty model applied to Danish twins. Twin Res. 2001;4:266-274. [PubMed] [Cited in This Article: ] |
| 25. | Sholtz RI, Rosenman RH, Brand RJ. The relationship of reported parental history to the incidence of coronary heart disease in the Western Collaborative Group Study. Am J Epidemiol. 1975;102:350-356. [PubMed] [Cited in This Article: ] |
| 26. | Colditz GA, Rimm EB, Giovannucci E, Stampfer MJ, Rosner B, Willett WC. A prospective study of parental history of myocardial infarction and coronary artery disease in men. Am J Cardiol. 1991;67:933-938. [PubMed] [Cited in This Article: ] |
| 27. | Colditz GA, Stampfer MJ, Willett WC, Rosner B, Speizer FE, Hennekens CH. A prospective study of parental history of myocardial infarction and coronary heart disease in women. Am J Epidemiol. 1986;123:48-58. [PubMed] [Cited in This Article: ] |
| 28. | Phillips AN, Shaper AG, Pocock SJ, Walker M. Parental death from heart disease and the risk of heart attack. Eur Heart J. 1988;9:243-251. [PubMed] [Cited in This Article: ] |
| 29. | Assmann G, Cullen P, Schulte H. Simple scoring scheme for calculating the risk of acute coronary events based on the 10-year follow-up of the prospective cardiovascular Münster (PROCAM) study. Circulation. 2002;105:310-315. [PubMed] [Cited in This Article: ] |
| 30. | Hopkins PN, Williams RR, Kuida H, Stults BM, Hunt SC, Barlow GK, Ash KO. Family history as an independent risk factor for incident coronary artery disease in a high-risk cohort in Utah. Am J Cardiol. 1988;62:703-707. [PubMed] [Cited in This Article: ] |
| 31. | Lloyd-Jones DM, Nam BH, D’Agostino RB, Levy D, Murabito JM, Wang TJ, Wilson PW, O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204-2211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 526] [Cited by in F6Publishing: 496] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 32. | Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937-952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7301] [Cited by in F6Publishing: 6998] [Article Influence: 349.9] [Reference Citation Analysis (1)] |
| 33. | Mangino M, Spector T. Understanding coronary artery disease using twin studies. Heart. 2013;99:373-375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 278] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 35. | Guella I, Rimoldi V, Asselta R, Ardissino D, Francolini M, Martinelli N, Girelli D, Peyvandi F, Tubaro M, Merlini PA. Association and functional analyses of MEF2A as a susceptibility gene for premature myocardial infarction and coronary artery disease. Circ Cardiovasc Genet. 2009;2:165-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Maiolino G, Colonna S, Zanchetta M, Pedon L, Seccia TM, Cesari M, Vigili de Kreutzenberg S, Avogaro A, Rossi GP. Exon 11 deletion in the myocyte enhancer factor (MEF)2A and early onset coronary artery disease gene in a Sicilian family. Eur J Cardiovasc Prev Rehabil. 2011;18:557-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 37. | Liu Y, Niu W, Wu Z, Su X, Chen Q, Lu L, Jin W. Variants in exon 11 of MEF2A gene and coronary artery disease: evidence from a case-control study, systematic review, and meta-analysis. PLoS One. 2012;7:e31406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Bhagavatula MR, Fan C, Shen GQ, Cassano J, Plow EF, Topol EJ, Wang Q. Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet. 2004;13:3181-3188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 39. | González P, García-Castro M, Reguero JR, Batalla A, Ordóñez AG, Palop RL, Lozano I, Montes M, Alvarez V, Coto E. The Pro279Leu variant in the transcription factor MEF2A is associated with myocardial infarction. J Med Genet. 2006;43:167-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Weng L, Kavaslar N, Ustaszewska A, Doelle H, Schackwitz W, Hébert S, Cohen JC, McPherson R, Pennacchio LA. Lack of MEF2A mutations in coronary artery disease. J Clin Invest. 2005;115:1016-1020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 41. | Lieb W, Mayer B, König IR, Borwitzky I, Götz A, Kain S, Hengstenberg C, Linsel-Nitschke P, Fischer M, Döring A. Lack of association between the MEF2A gene and myocardial infarction. Circulation. 2008;117:185-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 42. | Inanloo Rahatloo K, Davaran S, Elahi E. Lack of Association between the MEF2A Gene and Coronary Artery Disease in Iranian Families. Iran J Basic Med Sci. 2013;16:950-954. [PubMed] [Cited in This Article: ] |
| 43. | Dai DP, Zhou XY, Xiao Y, Xu F, Sun FC, Ji FS, Zhang ZX, Hu JH, Guo J, Zheng JD. Structural changes in exon 11 of MEF2A are not related to sporadic coronary artery disease in Han Chinese population. Eur J Clin Invest. 2010;40:669-677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 44. | Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278-1282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 452] [Cited by in F6Publishing: 459] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 45. | Wang H, Liu QJ, Chen MZ, Li L, Zhang K, Cheng GH, Ma L, Gong YQ. Association of common polymorphisms in the LRP6 gene with sporadic coronary artery disease in a Chinese population. Chin Med J (Engl). 2012;125:444-449. [PubMed] [Cited in This Article: ] |
| 46. | De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Sáez K, Henríquez JP, Zhao A. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer’s disease. Proc Natl Acad Sci USA. 2007;104:9434-9439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 216] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 47. | Sarzani R, Salvi F, Bordicchia M, Guerra F, Battistoni I, Pagliariccio G, Carbonari L, Dessì-Fulgheri P, Rappelli A. Carotid artery atherosclerosis in hypertensive patients with a functional LDL receptor-related protein 6 gene variant. Nutr Metab Cardiovasc Dis. 2011;21:150-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Xu S, Cheng J, Chen YN, Li K, Ma ZW, Cen JM, Liu X, Yang XL, Chen C, Xiong XD. The LRP6 rs2302685 polymorphism is associated with increased risk of myocardial infarction. Lipids Health Dis. 2014;13:94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 49. | Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 488] [Cited by in F6Publishing: 502] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 50. | Inanloorahatloo K, Zand Parsa AF, Huse K, Rasooli P, Davaran S, Platzer M, Fan JB, Amini S, Steemers F, Elahi E. Mutation in CYP27A1 identified in family with coronary artery disease. Eur J Med Genet. 2013;56:655-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 51. | Zurkinden L, Solcà C, Vögeli IA, Vogt B, Ackermann D, Erickson SK, Frey FJ, Sviridov D, Escher G. Effect of Cyp27A1 gene dosage on atherosclerosis development in ApoE-knockout mice. FASEB J. 2014;28:1198-1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 52. | InanlooRahatloo K, Parsa AF, Huse K, Rasooli P, Davaran S, Platzer M, Kramer M, Fan JB, Turk C, Amini S. Mutation in ST6GALNAC5 identified in family with coronary artery disease. Sci Rep. 2014;4:3595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Gopaul KP, Crook MA. Sialic acid: a novel marker of cardiovascular disease? Clin Biochem. 2006;39:667-681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 54. | Musunuru K, Kathiresan S. Genetics of coronary artery disease. Annu Rev Genomics Hum Genet. 2010;11:91-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 55. | British Heart Foundation. LDLR Database. Available from: http://www.ucl.ac.uk/ldlr/Current/summary.php?select_db=LDLR&show=sum. [Cited in This Article: ] |
| 56. | Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 806] [Cited by in F6Publishing: 790] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 57. | Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 804] [Cited by in F6Publishing: 847] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 58. | Kassner U, Wühle-Demuth M, Missala I, Humphries SE, Steinhagen-Thiessen E, Demuth I. Clinical utility gene card for: hyperlipoproteinemia, TYPE II. Eur J Hum Genet. 2014;22:Epub 2013 Nov 20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86:587-591. [PubMed] [Cited in This Article: ] |
| 60. | Pullinger CR, Hennessy LK, Chatterton JE, Liu W, Love JA, Mendel CM, Frost PH, Malloy MJ, Schumaker VN, Kane JP. Familial ligand-defective apolipoprotein B. Identification of a new mutation that decreases LDL receptor binding affinity. J Clin Invest. 1995;95:1225-1234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 146] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 61. | Thomas ER, Atanur SS, Norsworthy PJ, Encheva V, Snijders AP, Game L, Vandrovcova J, Siddiq A, Seed M, Soutar AK. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol Genet Genomic Med. 2013;1:155-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 62. | Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2100] [Cited by in F6Publishing: 2047] [Article Influence: 97.5] [Reference Citation Analysis (0)] |
| 63. | Abifadel M, Rabès JP, Devillers M, Munnich A, Erlich D, Junien C, Varret M, Boileau C. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Hum Mutat. 2009;30:520-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 64. | Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 781] [Cited by in F6Publishing: 830] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 65. | Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264-1272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2340] [Cited by in F6Publishing: 2222] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 66. | Arca M, Zuliani G, Wilund K, Campagna F, Fellin R, Bertolini S, Calandra S, Ricci G, Glorioso N, Maioli M. Autosomal recessive hypercholesterolaemia in Sardinia, Italy, and mutations in ARH: a clinical and molecular genetic analysis. Lancet. 2002;359:841-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 67. | Cohen JC, Kimmel M, Polanski A, Hobbs HH. Molecular mechanisms of autosomal recessive hypercholesterolemia. Curr Opin Lipidol. 2003;14:121-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 68. | Kiss RS, Kavaslar N, Okuhira K, Freeman MW, Walter S, Milne RW, McPherson R, Marcel YL. Genetic etiology of isolated low HDL syndrome: incidence and heterogeneity of efflux defects. Arterioscler Thromb Vasc Biol. 2007;27:1139-1145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 69. | Ordovas JM, Schaefer EJ, Salem D, Ward RH, Glueck CJ, Vergani C, Wilson PW, Karathanasis SK. Apolipoprotein A-I gene polymorphism associated with premature coronary artery disease and familial hypoalphalipoproteinemia. N Engl J Med. 1986;314:671-677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 161] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 70. | Yamakawa-Kobayashi K, Yanagi H, Fukayama H, Hirano C, Shimakura Y, Yamamoto N, Arinami T, Tsuchiya S, Hamaguchi H. Frequent occurrence of hypoalphalipoproteinemia due to mutant apolipoprotein A-I gene in the population: a population-based survey. Hum Mol Genet. 1999;8:331-336. [PubMed] [Cited in This Article: ] |
| 71. | Haase CL, Frikke-Schmidt R, Nordestgaard BG, Kateifides AK, Kardassis D, Nielsen LB, Andersen CB, Køber L, Johnsen AH, Grande P. Mutation in APOA1 predicts increased risk of ischaemic heart disease and total mortality without low HDL cholesterol levels. J Intern Med. 2011;270:136-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 72. | Haase CL, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Population-based resequencing of APOA1 in 10,330 individuals: spectrum of genetic variation, phenotype, and comparison with extreme phenotype approach. PLoS Genet. 2012;8:e1003063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Chiesa G, Sirtori CR. Apolipoprotein A-I(Milano): current perspectives. Curr Opin Lipidol. 2003;14:159-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 74. | Serfaty-Lacrosniere C, Civeira F, Lanzberg A, Isaia P, Berg J, Janus ED, Smith MP, Pritchard PH, Frohlich J, Lees RS. Homozygous Tangier disease and cardiovascular disease. Atherosclerosis. 1994;107:85-98. [PubMed] [Cited in This Article: ] |
| 75. | Asztalos BF, Brousseau ME, McNamara JR, Horvath KV, Roheim PS, Schaefer EJ. Subpopulations of high density lipoproteins in homozygous and heterozygous Tangier disease. Atherosclerosis. 2001;156:217-225. [PubMed] [Cited in This Article: ] |
| 76. | Brousseau ME, Bodzioch M, Schaefer EJ, Goldkamp AL, Kielar D, Probst M, Ordovas JM, Aslanidis C, Lackner KJ, Bloomfield Rubins H. Common variants in the gene encoding ATP-binding cassette transporter 1 in men with low HDL cholesterol levels and coronary heart disease. Atherosclerosis. 2001;154:607-611. [PubMed] [Cited in This Article: ] |
| 77. | Frikke-Schmidt R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis. 2010;208:305-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 78. | Cenarro A, Artieda M, Castillo S, Mozas P, Reyes G, Tejedor D, Alonso R, Mata P, Pocoví M, Civeira F. A common variant in the ABCA1 gene is associated with a lower risk for premature coronary heart disease in familial hypercholesterolaemia. J Med Genet. 2003;40:163-168. [PubMed] [Cited in This Article: ] |
| 79. | Holleboom AG, Kuivenhoven JA, Peelman F, Schimmel AW, Peter J, Defesche JC, Kastelein JJ, Hovingh GK, Stroes ES, Motazacker MM. High prevalence of mutations in LCAT in patients with low HDL cholesterol levels in The Netherlands: identification and characterization of eight novel mutations. Hum Mutat. 2011;32:1290-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 80. | Norum KR, Gjone E, Glomset JA. Familial lecithin: cholesterol acyltransferase deficiency including fish eye disease. The metabolic basis of inherited disease. New York: McGraw-Hill 1989; 1181-1194. [Cited in This Article: ] |
| 81. | Sarwar N, Danesh J, Eiriksdottir G, Sigurdsson G, Wareham N, Bingham S, Boekholdt SM, Khaw KT, Gudnason V. Triglycerides and the risk of coronary heart disease: 10,158 incident cases among 262,525 participants in 29 Western prospective studies. Circulation. 2007;115:450-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 940] [Cited by in F6Publishing: 1001] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 82. | Merkel M, Heeren J, Dudeck W, Rinninger F, Radner H, Breslow JL, Goldberg IJ, Zechner R, Greten H. Inactive lipoprotein lipase (LPL) alone increases selective cholesterol ester uptake in vivo, whereas in the presence of active LPL it also increases triglyceride hydrolysis and whole particle lipoprotein uptake. J Biol Chem. 2002;277:7405-7411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 76] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 83. | Evans D, Arzer J, Aberle J, Beil FU. Rare variants in the lipoprotein lipase (LPL) gene are common in hypertriglyceridemia but rare in Type III hyperlipidemia. Atherosclerosis. 2011;214:386-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 84. | Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1211] [Cited by in F6Publishing: 1227] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 85. | Connelly PW, Maguire GF, Little JA. Apolipoprotein CIISt. Michael. Familial apolipoprotein CII deficiency associated with premature vascular disease. J Clin Invest. 1987;80:1597-1606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 86. | Kawano M, Kodama K, Inadera H, Saito Y, Saito M, Yaginuma T, Kanazawa Y, Kawakami M. A case of apolipoprotein C-II deficiency with coronary artery disease. Clin Exp Med. 2002;2:29-31. [PubMed] [Cited in This Article: ] |
| 87. | Kaya Z, Niu DM, Yorulmaz A, Tekin A, Gürsel T. A novel mutation of ABCG5 gene in a Turkish boy with phytosterolemia presenting with macrotrombocytopenia and stomatocytosis. Pediatr Blood Cancer. 2014;61:1457-1459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 88. | Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335-1344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 615] [Cited by in F6Publishing: 570] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 89. | Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 212] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 90. | IBC 50K CAD Consortium. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 91. | Roberts R. Genetics of coronary artery disease: an update. Methodist Debakey Cardiovasc J. 2014;10:7-12. [PubMed] [Cited in This Article: ] |
| 92. | Lieb W, Vasan RS. Genetics of coronary artery disease. Circulation. 2013;128:1131-1138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 93. | Roberts R, Stewart AF. The genetics of coronary artery disease. Curr Opin Cardiol. 2012;27:221-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 94. | Roberts R, Stewart AF. Genes and coronary artery disease: where are we? J Am Coll Cardiol. 2012;60:1715-1721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 95. | Roberts R, Stewart AF. Genetics of coronary artery disease in the 21st century. Clin Cardiol. 2012;35:536-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 96. | Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551-1561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 202] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 97. | Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747-753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5794] [Cited by in F6Publishing: 5541] [Article Influence: 369.4] [Reference Citation Analysis (0)] |
| 98. | Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 99. | Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1245] [Cited by in F6Publishing: 1167] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 100. | Kitsios GD, Dahabreh IJ, Trikalinos TA, Schmid CH, Huggins GS, Kent DM. Heterogeneity of the phenotypic definition of coronary artery disease and its impact on genetic association studies. Circ Cardiovasc Genet. 2011;4:58-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 101. | Lin DY, Zeng D, Tang ZZ. Quantitative trait analysis in sequencing studies under trait-dependent sampling. Proc Natl Acad Sci USA. 2013;110:12247-12252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 102. | Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193-1198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1086] [Cited by in F6Publishing: 1017] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 103. | Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci USA. 2014;111:E455-E464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 456] [Cited by in F6Publishing: 415] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 104. | Hou L, Zhao H. A review of post-GWAS prioritization approaches. Front Genet. 2013;4:280. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 105. | Mäkinen VP, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segrè AV, Ghosh S, Vivar J. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 106. | Robinson MR, Wray NR, Visscher PM. Explaining additional genetic variation in complex traits. Trends Genet. 2014;30:124-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 107. | Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 321] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 108. | Ermann J, Glimcher LH. After GWAS: mice to the rescue? Curr Opin Immunol. 2012;24:564-570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 109. | Lusis AJ. Genetics of atherosclerosis. Trends Genet. 2012;28:267-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 110. | Terry DF, Wilcox M, McCormick MA, Lawler E, Perls TT. Cardiovascular advantages among the offspring of centenarians. J Gerontol A Biol Sci Med Sci. 2003;58:M425-M431. [PubMed] [Cited in This Article: ] |
| 111. | Hopkins PN. Molecular biology of atherosclerosis. Physiol Rev. 2013;93:1317-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 311] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 112. | Wang M, Lang X, Cui S, Zou L, Cao J, Wang S, Wu X. Quantitative assessment of the influence of paraoxonase 1 activity and coronary heart disease risk. DNA Cell Biol. 2012;31:975-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 113. | Zhao Y, Ma Y, Fang Y, Liu L, Wu S, Fu D, Wang X. Association between PON1 activity and coronary heart disease risk: a meta-analysis based on 43 studies. Mol Genet Metab. 2012;105:141-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 114. | Hong SH, Song J, Min WK, Kim JQ. Genetic variations of the paraoxonase gene in patients with coronary artery disease. Clin Biochem. 2001;34:475-481. [PubMed] [Cited in This Article: ] |
| 115. | Chen Q, Reis SE, Kammerer CM, McNamara DM, Holubkov R, Sharaf BL, Sopko G, Pauly DF, Merz CN, Kamboh MI. Association between the severity of angiographic coronary artery disease and paraoxonase gene polymorphisms in the National Heart, Lung, and Blood Institute-sponsored Women’s Ischemia Syndrome Evaluation (WISE) study. Am J Hum Genet. 2003;72:13-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 116. | Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, Fu X, Shao M, Brennan DM, Ellis SG. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265-1276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 412] [Cited by in F6Publishing: 409] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 117. | Tobin MD, Braund PS, Burton PR, Thompson JR, Steeds R, Channer K, Cheng S, Lindpaintner K, Samani NJ. Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J. 2004;25:459-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 110] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 118. | Shih DM, Xia YR, Wang XP, Miller E, Castellani LW, Subbanagounder G, Cheroutre H, Faull KF, Berliner JA, Witztum JL. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J Biol Chem. 2000;275:17527-17535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 305] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 119. | Zhang C, Peng W, Wang M, Zhu J, Zang Y, Shi W, Zhang J, Qin J. Studies on protective effects of human paraoxonases 1 and 3 on atherosclerosis in apolipoprotein E knockout mice. Gene Ther. 2010;17:626-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 120. | Tward A, Xia YR, Wang XP, Shi YS, Park C, Castellani LW, Lusis AJ, Shih DM. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation. 2002;106:484-490. [PubMed] [Cited in This Article: ] |
| 121. | Guns PJ, Van Assche T, Verreth W, Fransen P, Mackness B, Mackness M, Holvoet P, Bult H. Paraoxonase 1 gene transfer lowers vascular oxidative stress and improves vasomotor function in apolipoprotein E-deficient mice with pre-existing atherosclerosis. Br J Pharmacol. 2008;153:508-516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 122. | Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837-1847. [PubMed] [Cited in This Article: ] |
| 123. | Conroy RM, Pyörälä K, Fitzgerald AP, Sans S, Menotti A, De Backer G, De Bacquer D, Ducimetière P, Jousilahti P, Keil U. Estimation of ten-year risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J. 2003;24:987-1003. [PubMed] [Cited in This Article: ] |
| 124. | Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA. 2007;297:611-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1280] [Cited by in F6Publishing: 1216] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 125. | Marks D, Wonderling D, Thorogood M, Lambert H, Humphries SE, Neil HA. Cost effectiveness analysis of different approaches of screening for familial hypercholesterolaemia. BMJ. 2002;324:1303. [PubMed] [Cited in This Article: ] |
| 126. | Humphries SE, Ridker PM, Talmud PJ. Genetic testing for cardiovascular disease susceptibility: a useful clinical management tool or possible misinformation? Arterioscler Thromb Vasc Biol. 2004;24:628-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 127. | Janssens AC, Ioannidis JP, van Duijn CM, Little J, Khoury MJ. Strengthening the reporting of genetic risk prediction studies: the GRIPS statement. Genome Med. 2011;3:16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 128. | Janssens AC, Ioannidis JP, Bedrosian S, Boffetta P, Dolan SM, Dowling N, Fortier I, Freedman AN, Grimshaw JM, Gulcher J. Strengthening the reporting of Genetic RIsk Prediction Studies (GRIPS): explanation and elaboration. J Clin Epidemiol. 2011;64:e1-e22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 129. | Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 130. | Brautbar A, Ballantyne CM, Lawson K, Nambi V, Chambless L, Folsom AR, Willerson JT, Boerwinkle E. Impact of adding a single allele in the 9p21 locus to traditional risk factors on reclassification of coronary heart disease risk and implications for lipid-modifying therapy in the Atherosclerosis Risk in Communities study. Circ Cardiovasc Genet. 2009;2:279-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 131. | Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65-72. [PubMed] [Cited in This Article: ] |
| 132. | Davies RW, Dandona S, Stewart AF, Chen L, Ellis SG, Tang WH, Hazen SL, Roberts R, McPherson R, Wells GA. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet. 2010;3:468-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 133. | Gränsbo K, Almgren P, Sjögren M, Smith JG, Engström G, Hedblad B, Melander O. Chromosome 9p21 genetic variation explains 13% of cardiovascular disease incidence but does not improve risk prediction. J Intern Med. 2013;274:233-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 134. | Drenos F, Whittaker JC, Humphries SE. The use of meta-analysis risk estimates for candidate genes in combination to predict coronary heart disease risk. Ann Hum Genet. 2007;71:611-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 135. | van der Net JB, Janssens AC, Sijbrands EJ, Steyerberg EW. Value of genetic profiling for the prediction of coronary heart disease. Am Heart J. 2009;158:105-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 136. | Di Angelantonio E, Butterworth AS. Clinical utility of genetic variants for cardiovascular risk prediction: a futile exercise or insufficient data? Circ Cardiovasc Genet. 2012;5:387-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 137. | Sayols-Baixeras S, Lluís-Ganella C, Lucas G, Elosua R. Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Appl Clin Genet. 2014;7:15-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 138. | Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 222] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 139. | Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393-1400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 447] [Cited by in F6Publishing: 417] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 140. | Paynter NP, Chasman DI, Paré G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631-637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 335] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 141. | Qi L, Parast L, Cai T, Powers C, Gervino EV, Hauser TH, Hu FB, Doria A. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol. 2011;58:2675-2682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 142. | Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ, O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 143. | Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, Kontto J, Karvanen J, Willenborg C, Salomaa V. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7:e40922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 144. | Vaarhorst AA, Lu Y, Heijmans BT, Dollé ME, Böhringer S, Putter H, Imholz S, Merry AH, van Greevenbroek MM, Jukema JW. Literature-based genetic risk scores for coronary heart disease: the Cardiovascular Registry Maastricht (CAREMA) prospective cohort study. Circ Cardiovasc Genet. 2012;5:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 145. | Bolton JL, Stewart MC, Wilson JF, Anderson N, Price JF. Improvement in prediction of coronary heart disease risk over conventional risk factors using SNPs identified in genome-wide association studies. PLoS One. 2013;8:e57310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 146. | Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2261-2266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 147. | Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240-1249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 517] [Cited by in F6Publishing: 497] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 148. | Isaacs A, Willems SM, Bos D, Dehghan A, Hofman A, Ikram MA, Uitterlinden AG, Oostra BA, Franco OH, Witteman JC. Risk scores of common genetic variants for lipid levels influence atherosclerosis and incident coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2233-2239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 149. | Guella I, Duga S, Ardissino D, Merlini PA, Peyvandi F, Mannucci PM, Asselta R. Common variants in the haemostatic gene pathway contribute to risk of early-onset myocardial infarction in the Italian population. Thromb Haemost. 2011;106:655-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 150. | Tiret L, Bonnardeaux A, Poirier O, Ricard S, Marques-Vidal P, Evans A, Arveiler D, Luc G, Kee F, Ducimetière P. Synergistic effects of angiotensin-converting enzyme and angiotensin-II type 1 receptor gene polymorphisms on risk of myocardial infarction. Lancet. 1994;344:910-913. [PubMed] [Cited in This Article: ] |
| 151. | Erdmann J, Stark K, Esslinger UB, Rumpf PM, Koesling D, de Wit C, Kaiser FJ, Braunholz D, Medack A, Fischer M. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 152. | Grüntzig AR, Senning A, Siegenthaler WE. Nonoperative dilatation of coronary-artery stenosis: percutaneous transluminal coronary angioplasty. N Engl J Med. 1979;301:61-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1827] [Cited by in F6Publishing: 1526] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 153. | Fattori R, Piva T. Drug-eluting stents in vascular intervention. Lancet. 2003;361:247-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 274] [Cited by in F6Publishing: 235] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 154. | Kastrati A, Schömig A, Seyfarth M, Koch W, Elezi S, Böttiger C, Mehilli J, Schömig K, von Beckerath N. PlA polymorphism of platelet glycoprotein IIIa and risk of restenosis after coronary stent placement. Circulation. 1999;99:1005-1010. [PubMed] [Cited in This Article: ] |
| 155. | Jukema JW, Verschuren JJ, Ahmed TA, Quax PH. Restenosis after PCI. Part 1: pathophysiology and risk factors. Nat Rev Cardiol. 2012;9:53-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 208] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 156. | Virmani R, Kolodgie FD, Finn AV, Gold HK. Chapter 4, pathological anatomy of restenosis. Essentials of restenosis for the interventional cardiologist. Totowa, NJ: Hamana Press Inc 2007; 47-58. [Cited in This Article: ] |
| 157. | Sampietro ML, Trompet S, Verschuren JJ, Talens RP, Deelen J, Heijmans BT, de Winter RJ, Tio RA, Doevendans PA, Ganesh SK. A genome-wide association study identifies a region at chromosome 12 as a potential susceptibility locus for restenosis after percutaneous coronary intervention. Hum Mol Genet. 2011;20:4748-4757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 158. | Zargar S, Wakil S, Mobeirek AF, Al-Jafari AA. Involvement of ATP-binding cassette, subfamily A polymorphism with susceptibility to coronary artery disease. Biomed Rep. 2013;1:883-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 159. | Singh R, Smith E, Fathzadeh M, Liu W, Go GW, Subrahmanyan L, Faramarzi S, McKenna W, Mani A. Rare nonconservative LRP6 mutations are associated with metabolic syndrome. Hum Mutat. 2013;34:1221-1225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 160. | Abd El-Aziz TA, Mohamed RH, Hagrass HA. Increased risk of premature coronary artery disease in Egyptians with ABCA1 (R219K), CETP (TaqIB), and LCAT (4886C/T) genes polymorphism. J Clin Lipidol. 2014;8:381-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 161. | Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443-453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1578] [Cited by in F6Publishing: 1510] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 162. | Arvind P, Nair J, Jambunathan S, Kakkar VV, Shanker J. CELSR2-PSRC1-SORT1 gene expression and association with coronary artery disease and plasma lipid levels in an Asian Indian cohort. J Cardiol. 2014;64:339-346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 163. | Gorący J, Gorący I, Kaczmarczyk M, Parczewski M, Brykczyński M, Clark J, Safranow K, Ciechanowicz A. Low frequency haplotypes of E-selectin polymorphisms G2692A and C1901T give increased protection from coronary artery disease. Med Sci Monit. 2011;17:CR334-CR340. [PubMed] [Cited in This Article: ] |
| 164. | Qi L, Qi Q, Prudente S, Mendonca C, Andreozzi F, di Pietro N, Sturma M, Novelli V, Mannino GC, Formoso G. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA. 2013;310:821-828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 103] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 165. | Lio D, Candore G, Crivello A, Scola L, Colonna-Romano G, Cavallone L, Hoffmann E, Caruso M, Licastro F, Caldarera CM. Opposite effects of interleukin 10 common gene polymorphisms in cardiovascular diseases and in successful ageing: genetic background of male centenarians is protective against coronary heart disease. J Med Genet. 2004;41:790-794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 166. | Shen GQ, Girelli D, Li L, Olivieri O, Martinelli N, Chen Q, Topol EJ, Wang QK. Multi-allelic haplotype association identifies novel information different from single-SNP analysis: a new protective haplotype in the LRP8 gene is against familial and early-onset CAD and MI. Gene. 2013;521:78-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 167. | Teupser D, Baber R, Ceglarek U, Scholz M, Illig T, Gieger C, Holdt LM, Leichtle A, Greiser KH, Huster D. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3:331-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 168. | Wu Z, Lou Y, Jin W, Liu Y, Lu L, Lu G. The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma-2 gene (PPARγ2) is associated with increased risk of coronary artery disease: a meta-analysis. PLoS One. 2012;7:e53105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 169. | Galgani A, Valdes A, Erlich HA, Mano C, Cheng S, Petrone A, Sentinelli F, Berni A, Baroni MG, Buzzetti R. Homozygosity for the Ala allele of the PPARγ2 Pro12Ala polymorphism is associated with reduced risk of coronary artery disease. Dis Markers. 2010;29:259-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 8] [Reference Citation Analysis (0)] |
| 170. | Youssef SM, Mohamed N, Afef S, Khaldoun BH, Fadoua N, Fadhel NM, Naceur SM. A Pro 12 Ala substitution in the PPARγ2 polymorphism may decrease the number of diseased vessels and the severity of angiographic coronary artery. Coron Artery Dis. 2013;24:347-351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 171. | Wu Z, Lou Y, Jin W, Liu Y, Lu L, Lu G. The C161T polymorphism in the peroxisome proliferator-activated receptor gamma gene (PPARγ) is associated with risk of coronary artery disease: a meta-analysis. Mol Biol Rep. 2013;40:3101-3112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 172. | Bacci S, Menzaghi C, Ercolino T, Ma X, Rauseo A, Salvemini L, Vigna C, Fanelli R, Di Mario U, Doria A. The +276 G/T single nucleotide polymorphism of the adiponectin gene is associated with coronary artery disease in type 2 diabetic patients. Diabetes Care. 2004;27:2015-2020. [PubMed] [Cited in This Article: ] |
| 173. | Chiodini BD, Specchia C, Gori F, Barlera S, D’Orazio A, Pietri S, Crociati L, Nicolucci A, Franciosi M, Signorini S. Adiponectin gene polymorphisms and their effect on the risk of myocardial infarction and type 2 diabetes: an association study in an Italian population. Ther Adv Cardiovasc Dis. 2010;4:223-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 174. | Sun K, Li Y, Wei C, Tong Y, Zheng H, Guo Y. Recessive protective effect of ADIPOQ rs1501299 on cardiovascular diseases with type 2 diabetes: a meta-analysis. Mol Cell Endocrinol. 2012;349:162-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 175. | Saksi J, Ijäs P, Mäyränpää MI, Nuotio K, Isoviita PM, Tuimala J, Lehtonen-Smeds E, Kaste M, Jula A, Sinisalo J. Low-expression variant of fatty acid-binding protein 4 favors reduced manifestations of atherosclerotic disease and increased plaque stability. Circ Cardiovasc Genet. 2014;7:588-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 176. | Sutton BS, Crosslin DR, Shah SH, Nelson SC, Bassil A, Hale AB, Haynes C, Goldschmidt-Clermont PJ, Vance JM, Seo D. Comprehensive genetic analysis of the platelet activating factor acetylhydrolase (PLA2G7) gene and cardiovascular disease in case-control and family datasets. Hum Mol Genet. 2008;17:1318-1328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 177. | Wartiovaara U, Perola M, Mikkola H, Tötterman K, Savolainen V, Penttilä A, Grant PJ, Tikkanen MJ, Vartiainen E, Karhunen PJ. Association of FXIII Val34Leu with decreased risk of myocardial infarction in Finnish males. Atherosclerosis. 1999;142:295-300. [PubMed] [Cited in This Article: ] |
| 178. | Hancer VS, Diz-Kucukkaya R, Bilge AK, Ozben B, Oncul A, Ergen G, Nalcaci M. The association between factor XIII Val34Leu polymorphism and early myocardial infarction. Circ J. 2006;70:239-242. [PubMed] [Cited in This Article: ] |
| 179. | Chen X, Li S, Yang Y, Yang X, Liu Y, Liu Y, Hu W, Jin L, Wang X. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J Thromb Haemost. 2012;10:1508-1514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 180. | Liu X, Li Y, Wang L, Zhao Q, Lu X, Huang J, Fan Z, Gu D. The INSIG1 gene, not the INSIG2 gene, associated with coronary heart disease: tagSNPs and haplotype-based association study. The Beijing Atherosclerosis Study. Thromb Haemost. 2008;100:886-892. [PubMed] [Cited in This Article: ] |
| 181. | Tachmazidou I, Dedoussis G, Southam L, Farmaki AE, Ritchie GR, Xifara DK, Matchan A, Hatzikotoulas K, Rayner NW, Chen Y. A rare functional cardioprotective APOC3 variant has risen in frequency in distinct population isolates. Nat Commun. 2013;4:2872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 182. | Girelli D, Russo C, Ferraresi P, Olivieri O, Pinotti M, Friso S, Manzato F, Mazzucco A, Bernardi F, Corrocher R. Polymorphisms in the factor VII gene and the risk of myocardial infarction in patients with coronary artery disease. N Engl J Med. 2000;343:774-780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 166] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 183. | Niessner A, Marculescu R, Kvakan H, Haschemi A, Endler G, Weyand CM, Maurer G, Mannhalter C, Wojta J, Wagner O. Fractalkine receptor polymorphisms V2491 and T280M as genetic risk factors for restenosis. Thromb Haemost. 2005;94:1251-1256. [PubMed] [Cited in This Article: ] |
| 184. | Apostolakis S, Amanatidou V, Papadakis EG, Spandidos DA. Genetic diversity of CX3CR1 gene and coronary artery disease: new insights through a meta-analysis. Atherosclerosis. 2009;207:8-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 185. | Hashad IM, Abdel Rahman MF, Abdel-Maksoud SM, Amr KS, Effat LK, Shaban GM, Gad MZ. C242T polymorphism of NADPH oxidase p22phox gene reduces the risk of coronary artery disease in a random sample of Egyptian population. Mol Biol Rep. 2014;41:2281-2286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 186. | Xu Q, Yuan F, Shen X, Wen H, Li W, Cheng B, Wu J. Polymorphisms of C242T and A640G in CYBA gene and the risk of coronary artery disease: a meta-analysis. PLoS One. 2014;9:e84251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 187. | Mikkelsson J, Perola M, Penttilä A, Karhunen PJ. Platelet glycoprotein Ibalpha HPA-2 Met/VNTR B haplotype as a genetic predictor of myocardial infarction and sudden cardiac death. Circulation. 2001;104:876-880. [PubMed] [Cited in This Article: ] |
| 188. | Chung SL, Chiou KR, Charng MJ. 677TT polymorphism of methylenetetrahydrofolate reductase in combination with low serum vitamin B12 is associated with coronary in-stent restenosis. Catheter Cardiovasc Interv. 2006;67:349-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 189. | Monraats PS, Kurreeman FA, Pons D, Sewgobind VD, de Vries FR, Zwinderman AH, de Maat MP, Doevendans PA, de Winter RJ, Tio RA. Interleukin 10: a new risk marker for the development of restenosis after percutaneous coronary intervention. Genes Immun. 2007;8:44-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 190. | Yang Y, Guo SX, Yang ZY, Zhang T, Cao HM, Wang RX. Association between 1019C/T polymorphism of Connexin 37 gene and restenosis after coronary stenting. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013;30:456-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 191. | Guo SX, Yang ZY, Wang RX, Yang Y, Cao HM, Zhang T. Association between C1019T polymorphism of the connexin37 gene and coronary heart disease in patients with in-stent restenosis. Exp Ther Med. 2013;5:539-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 192. | Miranda-Malpica E, Martínez-Rios MA, Fragoso JM, Delgadillo-Rodríguez H, Rodríguez-Pérez JM, González-Quesada C, Martínez-Rodríguez N, Saldaña-Mendoza A, Peña-Duque MA, Vargas-Alarcón G. The interleukin 1B-511 polymorphism is associated with the risk of developing restenosis after coronary stenting in Mexican patients. Hum Immunol. 2008;69:116-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 193. | Shuvalova YA, Kaminnyi AI, Meshkov AN, Shirokov RO, Samko AN. Association between polymorphisms of eNOS and GPx-1 genes, activity of free-radical processes and in-stent restenosis. Mol Cell Biochem. 2012;370:241-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 194. | Verschuren JJ, Trompet S, Postmus I, Sampietro ML, Heijmans BT, Houwing-Duistermaat JJ, Slagboom PE, Jukema JW. Systematic testing of literature reported genetic variation associated with coronary restenosis: results of the GENDER Study. PLoS One. 2012;7:e42401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 195. | Rudez G, Pons D, Leebeek F, Monraats P, Schrevel M, Zwinderman A, de Winter R, Tio R, Doevendans P, Jukema W. Platelet receptor P2RY12 haplotypes predict restenosis after percutaneous coronary interventions. Hum Mutat. 2008;29:375-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 196. | Bienertova-Vasku J, Bienert P, Hlinomaz O, Vasku A. Common polymorphism +45T/G in adiponectin gene as potential modulator of in-stent restenosis development. Int J Cardiol. 2010;145:351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 197. | Vogiatzi K, Voudris V, Apostolakis S, Kochiadakis GE, Thomopoulou S, Zaravinos A, Spandidos DA. Genetic diversity of RANTES gene promoter and susceptibility to coronary artery disease and restenosis after percutaneous coronary intervention. Thromb Res. 2009;124:84-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 198. | Silvestre-Roig C, Fernández P, Mansego ML, van Tiel CM, Viana R, Anselmi CV, Condorelli G, de Winter RJ, Martín-Fuentes P, Solanas-Barca M. Genetic variants in CCNB1 associated with differential gene transcription and risk of coronary in-stent restenosis. Circ Cardiovasc Genet. 2014;7:59-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 199. | Shimada K, Miyauchi K, Mokuno H, Watanabe Y, Iwama Y, Shigekiyo M, Matsumoto M, Okazaki S, Tanimoto K, Kurata T. Promoter polymorphism in the CD14 gene and concentration of soluble CD14 in patients with in-stent restenosis after elective coronary stenting. Int J Cardiol. 2004;94:87-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 200. | Hamann L, Glaeser C, Schulz S, Gross M, Franke A, Nöthlings U, Schumann RR. A micro RNA-146a polymorphism is associated with coronary restenosis. Int J Immunogenet. 2014;41:393-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 201. | Monraats PS, Pires NM, Agema WR, Zwinderman AH, Schepers A, de Maat MP, Doevendans PA, de Winter RJ, Tio RA, Waltenberger J. Genetic inflammatory factors predict restenosis after percutaneous coronary interventions. Circulation. 2005;112:2417-2425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 202. | Falcone C, Emanuele E, Buzzi MP, Ballerini L, Repetto A, Canosi U, Mazzucchelli I, Schirinzi S, Sbarsi I, Boiocchi C. The -374T/A variant of the rage gene promoter is associated with clinical restenosis after coronary stent placement. Int J Immunopathol Pharmacol. 2007;20:771-777. [PubMed] [Cited in This Article: ] |
| 203. | Ferrero V, Ribichini F, Matullo G, Guarrera S, Carturan S, Vado A, Vassanelli C, Piazza A, Uslenghi E, Wijns W. Estrogen receptor-alpha polymorphisms and angiographic outcome after coronary artery stenting. Arterioscler Thromb Vasc Biol. 2003;23:2223-2228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 204. | Ortlepp JR, Hoffmann R, Killian A, Lauscher J, Merkelbach-Brese S, Hanrath P. The 4G/5G promotor polymorphism of the plasminogen activator inhibitor-1 gene and late lumen loss after coronary stent placement in smoking and nonsmoking patients. Clin Cardiol. 2001;24:585-591. [PubMed] [Cited in This Article: ] |
| 205. | Gomma AH, Elrayess MA, Knight CJ, Hawe E, Fox KM, Humphries SE. The endothelial nitric oxide synthase (Glu298Asp and -786T& gt; C) gene polymorphisms are associated with coronary in-stent restenosis. Eur Heart J. 2002;23:1955-1962. [PubMed] [Cited in This Article: ] |
| 206. | Suzuki T, Okumura K, Sone T, Kosokabe T, Tsuboi H, Kondo J, Mukawa H, Kamiya H, Tomida T, Imai H. The Glu298Asp polymorphism in endothelial nitric oxide synthase gene is associated with coronary in-stent restenosis. Int J Cardiol. 2002;86:71-76. [PubMed] [Cited in This Article: ] |
| 207. | Liu W, Liu Y, Jiang H, Ding X, Zhu R, Li B, Zhao Y. Plasma levels of interleukin 18, interleukin 10, and matrix metalloproteinase-9 and -137G/C polymorphism of interleukin 18 are associated with incidence of in-stent restenosis after percutaneous coronary intervention. Inflammation. 2013;36:1129-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 208. | Oguri M, Kato K, Hibino T, Yokoi K, Segawa T, Matsuo H, Watanabe S, Nozawa Y, Murohara T, Yamada Y. Identification of a polymorphism of UCP3 associated with recurrent in-stent restenosis of coronary arteries. Int J Mol Med. 2007;20:533-538. [PubMed] [Cited in This Article: ] |
| 209. | van Tiel CM, Bonta PI, Rittersma SZ, Beijk MA, Bradley EJ, Klous AM, Koch KT, Baas F, Jukema JW, Pons D. p27kip1-838C>A single nucleotide polymorphism is associated with restenosis risk after coronary stenting and modulates p27kip1 promoter activity. Circulation. 2009;120:669-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 210. | Monraats PS, Fang Y, Pons D, Pires NM, Pols HA, Zwinderman AH, de Maat MP, Doevendans PA, DeWinter RJ, Tio RA. Vitamin D receptor: a new risk marker for clinical restenosis after percutaneous coronary intervention. Expert Opin Ther Targets. 2010;14:243-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 211. | Shah SH, Hauser ER, Crosslin D, Wang L, Haynes C, Connelly J, Nelson S, Johnson J, Gadson S, Nelson CL. ALOX5AP variants are associated with in-stent restenosis after percutaneous coronary intervention. Atherosclerosis. 2008;201:148-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 212. | Kojima S, Iwai N, Tago N, Ono K, Ohmi K, Tsujimoto G, Takagi S, Miyazaki S, Nonogi H, Goto Y. p53Arg72Pro polymorphism of tumour suppressor protein is associated with luminal narrowing after coronary stent placement. Heart. 2004;90:1069-1070. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 213. | Amant C, Bauters C, Bodart JC, Lablanche JM, Grollier G, Danchin N, Hamon M, Richard F, Helbecque N, McFadden EP. D allele of the angiotensin I-converting enzyme is a major risk factor for restenosis after coronary stenting. Circulation. 1997;96:56-60. [PubMed] [Cited in This Article: ] |
| 214. | Ribichini F, Steffenino G, Dellavalle A, Matullo G, Colajanni E, Camilla T, Vado A, Benetton G, Uslenghi E, Piazza A. Plasma activity and insertion/deletion polymorphism of angiotensin I-converting enzyme: a major risk factor and a marker of risk for coronary stent restenosis. Circulation. 1998;97:147-154. [PubMed] [Cited in This Article: ] |
| 215. | Ribichini F, Ferrero V, Matullo G, Feola M, Vado A, Camilla T, Guarrera S, Carturan S, Vassanelli C, Uslenghi E. Association study of the I/D polymorphism and plasma angiotensin-converting enzyme (ACE) as risk factors for stent restenosis. Clin Sci (Lond). 2004;107:381-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 216. | Liu ZP, Huo Y, Li JP, Zhang Y, Xue L, Zhao CY, Hong XM, Huang AQ, Gao W. Polymorphism K469E of intercellular adhesion molecule-1 gene and restenosis after coronary stenting in Chinese patients. Chin Med J (Engl). 2004;117:172-175. [PubMed] [Cited in This Article: ] |
| 217. | Koch W, Böttiger C, Mehilli J, von Beckerath N, Neumann FJ, Schömig A, Kastrati A. Association of a CD18 gene polymorphism with a reduced risk of restenosis after coronary stenting. Am J Cardiol. 2001;88:1120-1124. [PubMed] [Cited in This Article: ] |
| 218. | Gulesserian T, Wenzel C, Endler G, Sunder-Plassmann R, Marsik C, Mannhalter C, Iordanova N, Gyöngyösi M, Wojta J, Mustafa S. Clinical restenosis after coronary stent implantation is associated with the heme oxygenase-1 gene promoter polymorphism and the heme oxygenase-1 +99G/C variant. Clin Chem. 2005;51:1661-1665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |