Published online Oct 26, 2019. doi: 10.4330/wjc.v11.i10.221
Peer-review started: March 12, 2019
First decision: June 13, 2019
Revised: June 17, 2019
Accepted: September 22, 2019
Article in press: September 22, 2019
Published online: October 26, 2019
The genetic cardiomyopathies are a group of disorders related by abnormal myocardial structure and function. Although individually rare, these diseases collectively represent a significant health burden since they usually develop early in life and are a major cause of morbidity and mortality amongst affected children. The heterogeneity and rarity of these disorders requires the use of an appropriate model system in order to characterize the mechanism of disease and develop useful therapeutics since standard drug trials are infeasible. A common approach to study human disease involves the use of animal models, especially rodents, but due to important biological and physiological differences, this model system may not recapitulate human disease. An alternative approach for studying the metabolic cardiomyopathies relies on the use of cellular models which have most frequently been immortalized cell lines or patient-derived fibroblasts. However, the recent introduction of induced pluripotent stem cells (iPSCs), which have the ability to differentiate into any cell type in the body, is of great interest and has the potential to revolutionize the study of rare diseases. In this paper we review the advantages and disadvantages of each model system by comparing their utility for the study of mitochondrial cardiomyopathy with a particular focus on the use of iPSCs in cardiovascular biology for the modeling of rare genetic or metabolic diseases.
Core tip: Several experimental model systems exist for the modeling of cardiomyopathies, including those caused by rare metabolic or mitochondrial diseases. We compare and contrast the cellular models that have been used to date to model several different mitochondrial disorders with a particular focus on the advantages and disadvantages of induced pluripotent stem cells.
- Citation: Jimenez-Tellez N, Greenway SC. Cellular models for human cardiomyopathy: What is the best option? World J Cardiol 2019; 11(10): 221-235
- URL: https://www.wjgnet.com/1949-8462/full/v11/i10/221.htm
- DOI: https://dx.doi.org/10.4330/wjc.v11.i10.221
The cardiomyopathies are defined as a group of diseases of the heart characterized by abnormal structure and function of the myocardium[1]. The cardiomyopathies have been classically grouped according to cardiac morphology with the major categories being: hypertrophic cardiomyopathy (HCM), restrictive cardiomyopathy, dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy and left ventricular non-compaction cardiomyopathy (LVNC)[2]. These groups can be further subdivided into genetic and acquired forms based on disease mechanism[2]. The genetic cardiomyopathies generally arise in childhood or early adulthood and include metabolic and monogenic diseases.
The inborn errors of metabolism (IEM) are a heterogeneous group of rare genetic diseases caused by defects in energy production or intermediary metabolism[3,4]. Within the pediatric cardiomyopathies, IEM affect between 5% and 26% of infants and children[5]. There are more than 40 different IEM that are associated with the development of cardiomyopathy[3]. The mitochondrial cardiomyopathies represent the largest subset and result from pathologic mutations in either mitochondrial or nuclear genes[6] that ultimately lead to dysfunction of the electron transport chain[7], the main supplier of cellular energy under aerobic conditions[8]. Since the heart is one of the most energy-demanding organ in the body[9], cardiomyopathies are found in 20%-40% of children with mitochondrial disease[10]. Given the early onset of these devastating multisystem diseases, research into disease mechanism and the identification of potential therapeutics is essential. However, the heterogeneity and rarity of the IEM and the mitochondrial cardiomyopathies preclude randomized clinical drug trials with standardized end-points. This makes disease modelling using animals or cells an essential component in the study of these diseases.
The use of animal models for research, with rodents in particular, continues to represent the most commonly used and successful approach in reductionist biology. However, despite its many successes, this methodology is still questioned because of ethical implications, the frequent inability to totally recapitulate human genetic variability[11] and the fact that important species-specific differences exist for many aspects of biology which complicate both the study of disease and the translation of therapies into human subjects[12]. For example, in cardiac research specifically, the use of rodent models may be limited due to substantial biological differences in the cardiovascular system between rodents and humans. Rodent hearts beat at considerably higher heart rates (200-300 beats per minute) than humans (60-100 beats per minute)[13] and the duration of the ventricular action potential is significantly shorter in rodents[14] compared to humans[15]. Additionally, cardiomyocytes differ in the proteins expressed in the myofilaments, which affects repolarization and calcium sensitivity[13]. One potential strategy to improve the utility of animal models is to create “humanized models” using genetic engineering[11] or engrafting animals with human cells or tissues and immune suppressing them to prevent rejection of the foreign material[16]. Although this type of model is useful for studying many conditions including cancer[17], infectious diseases[18] and liver disease[19], they have important limitations, especially in terms of time, cost and difficulties in creation and maintenance. Furthermore, these hybrid animal models are often not feasible for studying the heart and cardiovascular system.
The adult mammalian heart is composed of multiple cell types, including cardiomyocytes, fibroblasts, endothelial cells, vascular and perivascular cells. The composition of the heart varies greatly between species[20] but, in humans, cardiomyocytes are the dominant cell type by volume, encompassing 70%-85% of the total heart. Cardiomyocytes give rise to specialized cells such as atrial myocytes, ventricular myocytes and Purkinje cells[21] and are responsible for the generation of contractile force[22]. However, although the other cell types only account for a small portion of the overall total myocardial mass, they are essential for maintaining homeostasis by providing the extracellular matrix and intercellular communication networks necessary to ensure proper cardiac function[23]. Although cardiomyocytes may be dominant by volume, they are not the most abundant cells. Fibroblasts are actually the most common cell type in the heart and are vital for maintaining the structure, mechanical and electrical functions of the heart[24]. Cardiomyocytes and fibroblasts are the best-studied cardiac cells and, since both cell types have important functions in the heart, we would suggest that both need to be examined to fully comprehend the cardiomyopathies.
Cell culture, using cardiomyocytes, fibroblasts and other cardiac-related cells, represents another well-established system to study human biology, understand disease and assess response to therapeutics. Primary cells and immortalized cell lines derived from human tissues represent two commonly-used experimental models. Primary cells reflect disease biology most faithfully since they are directly isolated from the tissue of interest and they maintain the morphology, function and protein markers in the dish as they possessed in vivo, but they are relatively delicate cells that can be difficult to maintain in culture and have a finite lifespan with limited potential for expansion[25]. Immortalized cells are derived by altering cell-cycle check points or modifying telomerase activity and, although these cells don’t have a limited lifespan and are capable of sustained active proliferation, they frequently contain genetic aberrations that can accumulate over time and lead to cellular behaviours that are distinct from those demonstrated in vivo[26].
Another approach to model disease involves the use of patient-derived cells. These cells are obtained from an individual patient and therefore allow for the study of human disease in its original genetic context and also have important advantages over primary or immortalized cells. The two most commonly used patient-derived cell types used for research today are induced pluripotent stem cells (iPSCs) and fibroblasts. Given that the genetic background for an individual is preserved, the use of these patient-specific cells represents perhaps the best tool to realize personalized medicine[27]. Personalized medicine refers to a health care approach which recognizes each person’s distinct genetic, clinical and environmental history[28]. Personalized medicine ideally adapts therapeutics in order to ensure the best response and safety for the treatment of specific diseases with an individualized approach[29]. Using patient-specific cells can help realize this vision by helping researchers identify and understand individual differences.
In conclusion, there are important differences between model systems (Table 1), with advantages and disadvantages that are often dependent on the condition being studied. In reality, a combination of models enabling both in vivo and in vitro studies is often required. In this paper, our main focus will be to discuss and compare the different cell types which could be useful for studying genetic cardiomyopathies as an alternative to primary cardiac cells. We will illustrate our discussion with examples of mitochondrial cardiomyopathies that have been studied using different cellular models.
| Properties | Animal | Cellular |
| Maintain genetic background | No | Yes |
| Cost of maintenance | Expensive | Less Expensive |
| Ease of maintenance | Simple | Difficult |
| Time required | +++ | + |
| Drug effects | Potentially not translatable | Translatable |
| Study of paracrine effects | Yes | No |
| Study of circulatory effects | Yes | No |
Immortalized cells are defined as cells whose proliferative capacity has been enhanced using different methods[30]. There are a variety of established approaches to immortalize cell lines including the introduction of oncogenes[31-33], viral transformation[34,35], the inactivation of tumor suppressor genes[36,37] or the inactivation of telomere-controlled senescence[38]. The establishment of immortalized cell lines has helped the scientific community to study different biological and molecular events[26], although, this approach has been questioned since these immortalized cells differ significantly from cells with an intact cell cycle control and they are more similar to malignant cells in many respects. Therefore, the results obtained with these cells can potentially be misleading if these differences are not considered[39]. However, the use of immortalized cells still remains one of the most popular models for the study of disease.
Immortalized cells have been used to study two inherited diseases caused by point mutations in mitochondrial DNA (mtDNA), mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS), and myoclonic epilepsy and ragged-red fibres (MERRF). In both diseases, an alteration in the post-transcriptional modification of a uridine located in an essential position of specific mitochondrial tRNAs, causes oxidative phosphorylation impairment that leads to the inability to generate sufficient ATP to meet the energy demands of the cell[40]. These mitochondrial disorders can be caused by mutations in several genes but, in this example, the immortalized cells were used to model the effect of an A>G transition at nucleotide 3243 in the tRNALeu gene causing MELAS[41] and a A>G change in the tRNALys gene at position 8344 causing MERRF[42]. Two different studies recapitulated these diseases using cybrid cells[43,44]. Cytoplasmic hybrid cells (cybrid) are created using a recipient cell line called rho-zero cells, whose mtDNA has been depleted but the nuclear DNA remains intact and a donor cell which provides mtDNA to the union[45]. This approach has the advantage of being able to isolate mtDNA from a donor patient with a specific mtDNA mutation, allowing for the study of the pathology in an immortalized cell line.
Another rare human disorder, Barth syndrome (BTHS) was studied using immortalized cell lines. BTHS is an X-linked recessive disorder characterized by early-onset cardiomyopathy (usually LVNC or DCM), skeletal muscle weakness and neutropenia related to abnormal mitochondrial structure[46]. Disease severity is highly variable, with patients ranging from being asymptomatic to having severe cardiomyopathy and end-stage heart failure[47]. Studies have shown that BTHS is caused by loss-of-function mutations in the tafazzin (TAZ) gene[48]. TAZ is a phospholipid transacylase located in the inner mitochondrial membrane and is responsible for remodeling of the phospholipid cardiolipin[49] which is an essential component of the mitochondrial membrane[50,51]. The TAZ gene consists of 11 different exons[52] and mutations have been identified in each exon, primarily missense mutations, although small insertions and deletions have also been found[53].
To study BTHS, the authors used a myoblast cell line (C2C12) derived from mouse skeletal myoblast cells, which is commonly used as a model of disease in mammals for skeletal muscle disorders and myopathies[54,55]. The authors designed a stable TAZ knockout (KO) using clustered regularly interspaced short palindromic repeats (CRISPR) technology to target exon 3 in mouse TAZ and cloned it into a plasmid together with the Cas9 nuclease and co-transfected into the cells with a plasmid that allowed for selection with puromycin[56]. With the introduction of the plasmids into the cell, the guide RNA binds to exogenous exon 3, and this binding is recognized by the nuclease, which performs the cutting of the gene, disrupting it. The clone whose genomic TAZ DNA band was fragmented into three pieces was the chosen one to be the model of the disease. According to the authors, this model served to recapitulate BTHS, being consistent with other previous models, showing mitochondrial defects such as accumulation of monolyso-cardiolipin, impaired mitochondrial respiration and increased mitochondrial ROS species[56].
Although these studies have used different immortalized cell models, these might not be the best tool to recapitulate the diseases with accuracy. First of all, these cells are derived either from tumors or from the immortalization of other cell times where the cell cycle or the telomerase activity is compromised, therefore, these cells do not resemble normal cell lines in terms of replication and lifespan and, consequently, this can cause genetic and phenotypic variation over time leading to create heterogeneity in the same cell line[57]. Secondly, these cell lines, like all cell lines are vulnerable to contamination (e.g., Mycoplasma) which can remain undetected and modify cell behaviour and gene expression[58]. Finally, the use of cellular models generated by using techniques that knockout a gene in particular in a cell line, might not be sufficient to recapitulate the entire spectrum of disease since additional genetic modifiers are not reproduced.
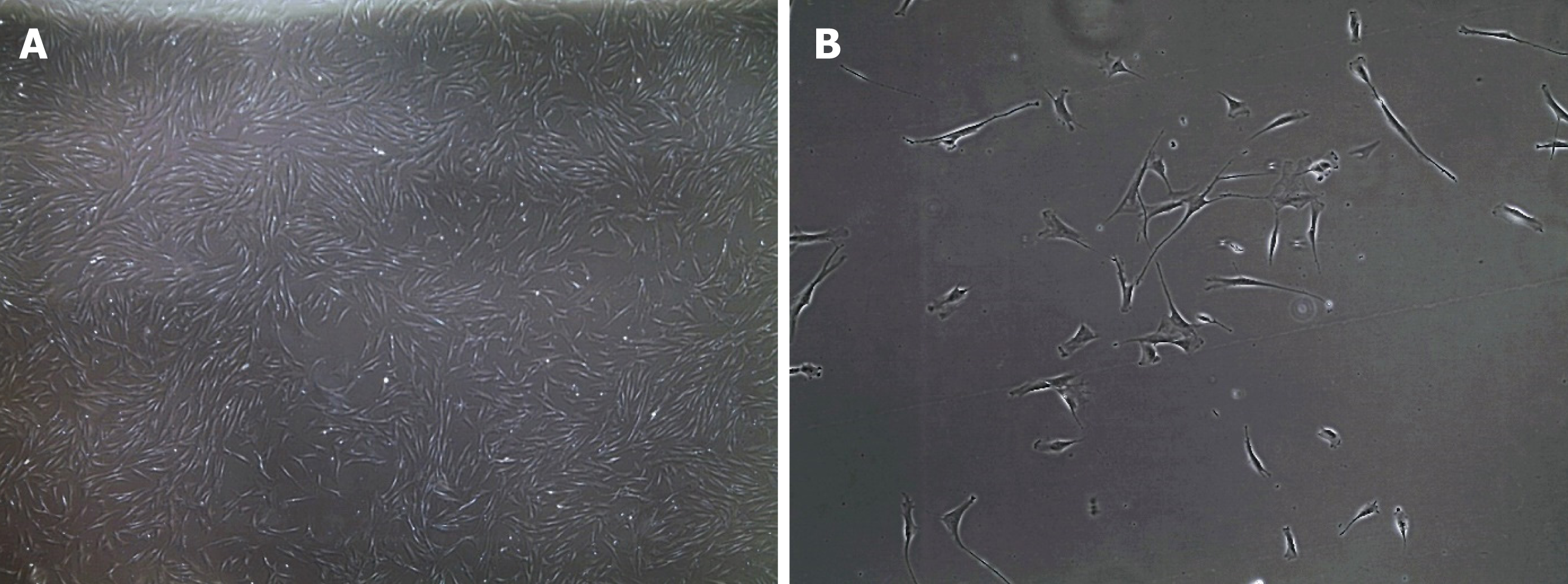
Fibroblasts are the major stromal cell-type present in connective tissue and are characterized by a flattened and elongated shape with a central nucleus[59] (Figure 1). They are derived from mesenchymal precursors and are part of a heterogenous collection of cells widely distributed over the body. Fibroblasts play an important role in connective tissue by producing extracellular matrix compounds, principally collagen type I and III. Fibroblasts not only have a structural role but they are able to repair damaged tissue by migrating to the site of injury and rapidly proliferating to restore the wounded area[60]. This proliferation potential explains why fibroblasts are so widely used and why they grow in vitro very easily[61]. In addition to their growth-related properties, fibroblasts are also increasingly recognized as an important contributor to cardiac biology through cell-cell signalling and physical interactions[62,63]. Unfortunately, fibroblasts have distinct electrophysiological properties and these cells are not electrically excitable despite the presence of multiple ion channels, including potassium and sodium channels[64]. Fibroblasts also lack a specific cell surface marker that distinguishes them from other cell types[65]. However, they can be isolated from a skin biopsy and grown in culture[66] but they do have a limited lifespan[67], so their use to study function, structure and disease mechanism is limited to cells that have not undergone an excessive (< 20) number or cell divisions or passages[66].
Fibroblasts have also been used to study MELAS and MERRF. This study demonstrated that the tRNA point mutations did not modify the number of normal mitochondria but there were important differences found regarding the number of secondary lysosomes and residual bodies in both diseases compared to the control cells[68]. Furthermore, in both diseases, there was impaired respiratory enzyme activity which decreased mitochondrial respiration rate and membrane potential and impacted cell viability due to the inability to synthesize enough ATP to meet the energy requirements of the cell[68]. Even though the cell types affected by MELAS and MERRF in humans are mainly neurons and myocytes[69,70], the easily obtainable skin fibroblasts were sufficient to provide a helpful model to understand some of the mechanisms by which these cell types are compromised. Fibroblasts were also used in BTHS to help understand the molecular basis of the disease. As previously mentioned, diverse mutations have been found in each exon of TAZ, however, there is no clear correlation between the gene mutation type and the different patient phenotypes[71]. The authors used fibroblasts from pediatric patients to correlate the severity of the disease with cellular lipid abnormalities and found that there was abnormal composition of cardiolipin, phosphatidyl-choline and phosphatidylethanolamine[72]. In this study fibroblasts allowed the distinct lipid composition for each patient to be characterized, which enabled insight into the phenotypic complexity of the disease[72].
Although all these studies successfully used fibroblasts to analyze different mitochondrial cardiomyopathies, all studies had to work within the limitation of fibroblast passage number. The passage number refers to the number of times that the cell can undergo cell division and replication. Studies have shown that, with every passage, the number of mitochondria decreases and that there are changes in the structure of these organelles[73]. If not recognized and controlled for, these changes have the potential to mislead researchers into making false conclusions regarding mitochondrial morphology and function.
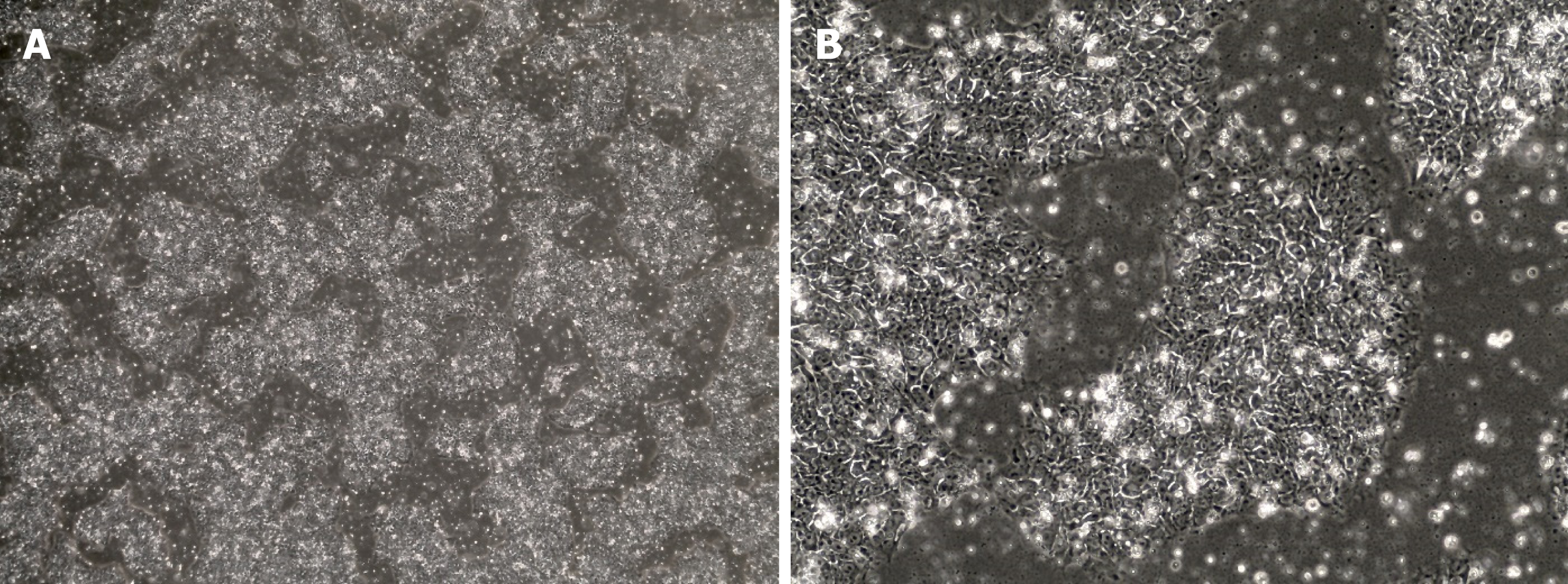
iPSCs were first created in 2006 after Shinya Yamanaka successfully reprogrammed adult mouse fibroblasts into iPSCs by introducing the pluripotency factors Oct3/4 (Octamer binding transcription factor 3/4), Sox2 (sex determining region Y)-box 2), c-Myc and Kfl4 (Kruppel Like Factor-4) under embryonic stem cells (ESC) conditions[74]. ESCs are derived from the inner cell mass of mammalian blastocysts and possess self-renewal capacity, the ability to grow with an unlimited lifespan and the ability to maintain pluripotency and differentiate into every cell type of the three germ layers[75,76]. The iPSCs created with these “Yamanaka factors” showed the morphology (Figure 2), proliferative properties and gene expression associated with pluripotency in ESCs[74] but, importantly, did not have to be derived from discarded human embryos. Currently, iPSCs can be created from a variety of mature, differentiated cells most commonly fibroblasts and peripheral blood mononuclear cells[77].
There are several technical approaches for the delivery of the four critical pluripotency factors necessary for cellular reprogramming to occur[78]. There are integrating methods that include retroviral transduction[74], lentiviral delivery[79] and non-integrative methods such as adenoviral transduction[80], plasmid DNA (episomal) transfer[81], lox p lentivirus delivery[82], Sendai virus delivery[83], piggyBAC transposon[84], protein-mediated (polyarginine-tagged polypeptide)[85] and modified synthetic mRNA[86] (Table 2). Each methodology has its advantages and disadvantages[87-89] and the choice of delivery vector can have important implications in downstream applications and, therefore, needs to be considered carefully.
| Method | Advantages | Disadvantages |
| Retroviral transduction | Efficient, validated for multiple cell types, easy | Transgene integration |
| Lentiviral delivery | Very efficient | Transgene integration |
| Adenoviral transduction | Does not integrate | Low efficiency, only validated for fibroblasts |
| Plasmid DNA transfer (episomal) | Good efficiency, does not integrate, able to replicate autonomously, validated for multiple cell types | Low efficiency in fibroblast reprogramming |
| Lox p lentivirus delivery | High efficiency, excision of the integrated sequence, gene expression profile closer to hES cells | Genomic instability and genome rearrangements and loxP site remains integrated |
| Sendai virus | Efficient, does not integrate, validated for multiple cell types | Cost if purchased commercially or challenging if generated by a laboratory |
| PiggyBAC transposon | Efficient, precise and efficient self-excision, does not remain integrated | Published work only in fibroblasts, licensing patent issues, pBt gene may remain active post-transposition |
| Polyarginine tagged polypeptide | Does not integrate | Low efficiency, time-consuming, technically challenging and work only on fibroblasts |
| RNA modified synthetic mRNA | Very efficient, does not integrate, factor available commercially | Cost if purchased commercially or challenging if generated by a laboratory and work only on fibroblasts |
Once created, iPSCs have significant advantages compared to other cell types as a model of disease. Since they possess the ability to self-renew, there is no concern about how many passages the cells can tolerate and these cells can be relatively easily expanded in vitro and be used for many experiments[90]. Furthermore, since they can be differentiated into mostly every cell type[91], researchers can generate patient- disease- and tissue-specific cells for the disease of interest.
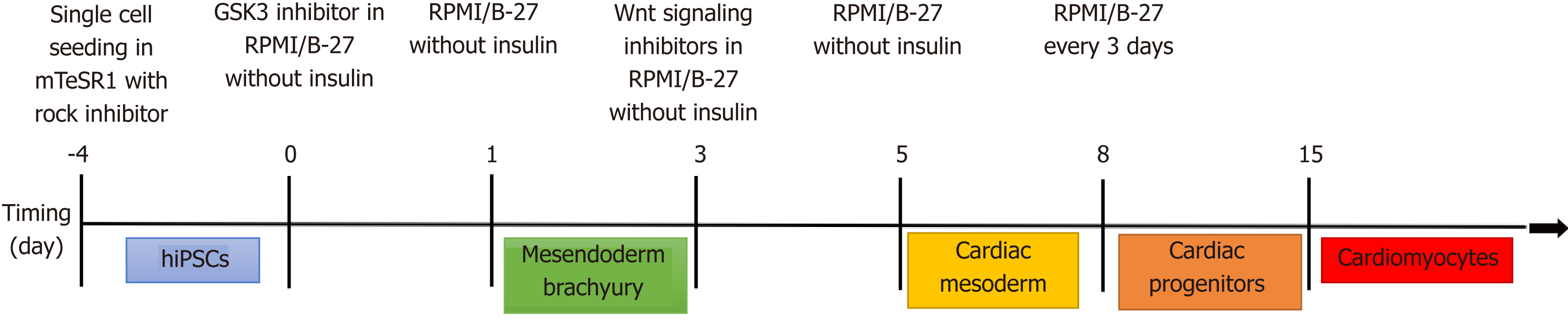
Most applications using iPSCs to study human heart disease have differentiated them into beating cardiomyocytes[92] although one group (discussed later) took a rather unique approach and differentiated the iPSCs back into fibroblasts[93]. There are several different published and commercial methods to differentiate iPSCs into cardiomyocytes all of which are generally based on the signaling factors that are part of the developmental pathway of cardiomyocytes in vivo[94-96] (Figure 3).
Although the ability to generate patient- and disease-specific beating cardiomyocytes is a powerful tool for the study of individual cardiomyopathies[97], the cardiomyocytes that are generated using current methods do have some limitations. First of all, following differentiation, the final population of cardiomyocytes are not completely homogeneous. Differentiated cells contain a mixture of atrial, ventricular and Purkinje cell-types with variable functional properties[98]. If a homogeneous population is desired, it may be necessary to select for the cellular subpopulation of interest using sorting techniques based on surface marker expression[99] or genetic selection[100] which further complicates the process requiring additional time and expense and exposes the cells to additional handling and stresses which they may not survive. Furthermore, for some cell types, e.g. ventricular myocytes, unique cell surface markers do not exist[101]. Another issue is that the cardiomyocytes obtained using current differentiation strategies have a phenotype resembling fetal cells in terms of structure, molecular markers and metabolism[102]. This lack of maturity can require additional steps (which are not fully established or reliably reproducible at this time) or additional time in culture to obtain a more adult-like cardiomyocyte population[103] Several methods to stimulate the maturation of iPSC-derived cardiomyocytes have been published based upon electrical[104], mechanical[105], chemical stimulation[106] or matrix modification[107]. This is currently an area of active investigation and future advances and improvements are certain which will further enhance the utility of iPSC-CMs for the study of genetic cardiomyopathies. However, even with these functional limitations of derived cells, they have been helpful for scientists seeking insight into cardiac biology and disease[108,109].
Primary fibroblasts from a patient with MELAS were reprogrammed into iPSCs using a retroviral approach in order to establish a novel disease model[110]. As standard practice, the differentiation capacities of the iPSCs were tested using a teratoma formation assay to demonstrate that the cells were capable of generating all germ layers and immunocytochemistry for the pluripotency markers Oct-4 and SSEA-4 was performed to confirm pluripotency. Tissues in MELAS patients can vary in the levels of abnormal mitochondria (heteroplasmy)[111] so the researchers assessed this in patient cells using quantitative real-time PCR to measure mutation ratios and mtDNA copy number. They found that different fibroblast lines had different levels of heteroplasmy ranging from < 5% to 95%. They then demonstrated that those fibroblasts with lower levels of heteroplasmy showed increased heteroplasmy after several passages while those with higher levels did not vary significantly after multiple passages. There were also variations with regards to mtDNA copy number after each passage. This data suggests that the mitochondrial abnormalities in patient fibroblasts can change over time in culture. However, because of their importance in cardiac biology, the authors still wanted to study MELAS. Therefore, the MELAS iPSCs were differentiated back into fibroblasts but, because of the unique self-renewing properties of iPSCs, the authors could overcome passage-associated changes in the mitochondria. In the fibroblasts derived from patient iPSCs, levels of heteroplasmy were found to be similar to the iPSCs from which they were differentiated. These iPSC-derived fibroblasts were then characterized with regards to the enzymatic activities of the mitochondrial respiratory complexes and compared to primary skin fibroblasts. These studies revealed that the iPSC-derived cells recapitulated the disease phenotype and did not demonstrate altered levels of heteroplasmy in culture and therefore represent a unique and novel in vitro model of MELAS[110].
MERRF has also been studied using retrovirus-reprogrammed iPSCs. In this study, they generated iPSCs from patient dermal fibroblasts. After reprogramming the fibroblasts using OCT4, SOX2, KLF4, and GLIS1 delivered into the cells, they differentiated the resulting iPSCs into the two different cell types most involved in the disease, cardiomyocytes (iPSC-CMs)[112] and neural progenitor cells (iPSC-NPCs). When they tested all three cell types, they found that all MERRF patient-derived cells (iPSCs, iPSC-CMs and iPSC-NPCs) had reduced oxygen consumption, elevated reactive oxygen species (ROS), reduced growth and fragmented mitochondria. The cellular phenotype correlated with the molecular mechanism of the disease, allowing iPSCs and iPSC-derived cells to serve as a model for the disease[93].
Differentiated iPSCs have also been used in the study of BTHS. The cells of two unrelated patients were reprogrammed using either retroviral[113] or modified RNA approaches[114]. These two patients had different mutations in TAZ, one having a frameshift mutation and the other a missense mutation. After the generation of the iPSCs, they differentiated them into cardiomyocytes that they then used to create tissue layers and a heart-on-chip model[115]. The iPSC-CMs showed abnormalities in cardiolipin processing, sarcomere assembly, myocardial contraction, ROS production and cardiomyocyte functioning, correlating with the abnormalities and cardiac dysfunction observed in patients, demonstrating again that is possible to use an in vitro model to provide insight into human disease and test potential therapeutics[116].
iPSC-CMs have also been used to study other cardiomyopathies. For example, iPSC-CMs have also been used to understand the pathological effects caused by the reduced expression of frataxin (FXN) in Friedreich ataxia (FA). This neurode-generative disease is caused by the expansion of a short tandem repeat (GAA) in the FXN gene, which can result in transcriptional silencing[117] and therefore, the development of HCM[118] which is an important component of the disease phenotype but its development is not understood. In this study, the researchers generated iPSCs from three patients using an episomal reprogramming approach and then differentiated the resulting iPSCs into cardiomyocytes[119]. Analysis of the iPSC-CMs showed that these cells had an increased beating rate which was related to a defect in calcium handling. Therefore, these cells revealed novel biology that could potentially contribute to the future development of treatment for this disease[120]. It is important to note that this cellular phenotype could arguably not have been accomplished with any other cell type.
The DCM with ataxia syndrome (DCMA) is an autosomal recessive disorder caused by mutation in DNAJC19 and is characterized by 39% mortality[121] during early childhood due to severe heart failure[122]. DCMA has been related to BTHS due to the presence of metabolic abnormalities (i.e. production of 3-methylglutaconic acid) and abnormal mitochondria are thought to be responsible for heart failure[123]. Rohani et al[123] successfully established four patient iPSC lines that have been differentiated into CMs expressing cardiac-specific markers and this will allow for the study of four unique patient cell lines. This disease still needs to be further characterized but the use of iPSC-CMs derived from patients looks promising as a cellular model to provide a better understanding of the disease.
Finally, iPSC-CMs have also been used to study familial HCM, characterized by thickened left ventricular walls, myofiber disarrays and myocardial fibrosis that often results in arrhythmias[124]. This can be caused by different mutation in at least 11 different genes which encode sarcomeric proteins[125]. In this study, the authors generated iPSC-CMs derived from an HCM patient that had a single missense mutation in the β-myosin heavy chain (MYH7) gene. Whole transcriptional analysis of these iPSC-CMs provided useful insights into the disease, revealing important signaling pathways implicated in the pathogenicity of HCM[126].
As we have described, both fibroblasts and iPSCs have been used to model genetic cardiomyopathies and both cell types have important advantages and disadvantages (Table 3). The characteristics of a specific cell type and the disease being studied may have an important influence on the researcher’s choice of cellular model and, in some situations, the study of both fibroblasts and iPSCs may be complementary. For instance, in a disease in which the interaction between cardiomyocytes and fibroblast plays a role in the development of the pathogenesis, for example in cardiac fibrosis and arrhythmias[127], the study of both cell types would likely be beneficial.
| Properties | Fibroblasts | iPSCs |
| Proliferation capacity | + | ++ |
| Self-renewal | No | Yes |
| Longevity | Limited | Unlimited |
| Differentiation | No | Yes |
| Metabolism | Quiescent | Energetic |
| Acquisition | Easy | Difficult |
| Cost | + | +++ |
| Ease of maintenance | Simple | Difficult |
| Necessary expertise | Low | High |
| Disease modeling | + | ++ |
| Structure | Single elongated cells | Round colonies/beating CM sheets |
| Maturation | Not applicable | Required for CM |
In order to solve the lifespan problem with primary cells such as fibroblasts, reversible immortalization could be performed to increase the number of passages and limit the risk for the development of aberrations in the genome[128]. In one study, this reversible immortalization was performed in primary neonatal rat cardio-myocytes using lentiviral transduction with either simian virus 40 large T antigen (TAg) or Bmi-1 together with the human telomerase reverse transcriptase (hTERT). After the cells were expanded, the introduced genes were removed using an adenoviral vector expressing Cre recombinase. The transduction of Bm1-1/hTERT into the primary cardiomyocytes successfully immortalized the cells and they maintained the expected cell morphology and presence of contact inhibition, suggesting that the cells had not become aberrant during the immortalization process[129]. This technique is an example of how genetic engineering could be used to overcome some of the limitations of cell biology which may be useful to researchers seeking to study a particular cell type.
Although patient-derived iPSCs and the differentiated cells that are created are excellent models of disease, the generation of appropriate controls is essential since they will help to define the abnormal phenotype. For some diseases that are enriched in specific populations with a unique genetic background, for example, DCMA, which is highly prevalent in the Hutterite population of southern Alberta[130], there is a need for controls who also have the same genetic background. The Hutterites are an isolated and genetically-closed population descended from a limited number of European ancestors with a communal religious lifestyle[131]. CRISPR/Cas9[132] can be used to repair the DNA mutation in patient cells to create isogenic controls[133] that are genetically identical except for a single genetic mutation background[134].
Cellular models represent an important tool for investigating rare human diseases including the genetic cardiomyopathies. Generic immortalized cells are the most commonly used cell model as they are the easiest to handle in terms of proliferation capacity, growth rate and low maintenance and can be easily genetically manipulated. Conversely, obtaining cells from individual patients allows the study of inter-individual differences and the important role of genetic modifiers in shaping disease phenotype and increases the possibility of developing personalized therapeutics. Certainly, in vitro models have some significant limitations but, in many cases, can provide a model that is otherwise not available. Particularly for cells differentiated from iPSCs, it is true that further research is necessary to optimize these cells but the potential for the development of an accurate and personalized cellular model is very promising for those diseases where conventional cells and animal models are limited.
Manuscript source: Invited manuscript
Specialty type: Cardiac and cardiovascular systems
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nurzynska D, Vidal-Perez R S-Editor: Gong ZM L-Editor: A E-Editor: Xing YX
| 1. | Sisakian H. Cardiomyopathies: Evolution of pathogenesis concepts and potential for new therapies. World J Cardiol. 2014;6:478-494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (3)] |
| 2. | Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB; American Heart Association; Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807-1816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2284] [Cited by in F6Publishing: 2118] [Article Influence: 117.7] [Reference Citation Analysis (0)] |
| 3. | Cox GF. Diagnostic Approaches to Pediatric Cardiomyopathy of Metabolic Genetic Etiologies and Their Relation to Therapy. Prog Pediatr Cardiol. 2007;24:15-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Mak CM, Lee HC, Chan AY, Lam CW. Inborn errors of metabolism and expanded newborn screening: review and update. Crit Rev Clin Lab Sci. 2013;50:142-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 5. | Byers SL, Ficicioglu C. Infant with cardiomyopathy: When to suspect inborn errors of metabolism? World J Cardiol. 2014;6:1149-1155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 6. | Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders--past, present and future. Biochim Biophys Acta. 2004;1659:115-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 219] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 7. | El-Hattab AW, Scaglia F. Mitochondrial Cardiomyopathies. Front Cardiovasc Med. 2016;3:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Munnich A, Rötig A, Rio M. Defects of the Respiratory Chain. Inborn Metabolic Diseases. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012; 223-238. [DOI] [Cited in This Article: ] |
| 9. | Chinnery PF, Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. Mitochondrial Disorders Overview 2014. [PubMed] [Cited in This Article: ] |
| 10. | Holmgren D, Wåhlander H, Eriksson BO, Oldfors A, Holme E, Tulinius M. Cardiomyopathy in children with mitochondrial disease; clinical course and cardiological findings. Eur Heart J. 2003;24:280-288. [PubMed] [Cited in This Article: ] |
| 11. | Ericsson AC, Crim MJ, Franklin CL. A brief history of animal modeling. Mo Med. 2013;110:201-205. [PubMed] [Cited in This Article: ] |
| 12. | Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12:786-798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 672] [Cited by in F6Publishing: 667] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 13. | Milani-Nejad N, Janssen PM. Small and large animal models in cardiac contraction research: advantages and disadvantages. Pharmacol Ther. 2014;141:235-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 295] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 14. | Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends Cardiovasc Med. 2004;14:83-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 129] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 15. | Glukhov AV, Fedorov VV, Lou Q, Ravikumar VK, Kalish PW, Schuessler RB, Moazami N, Efimov IR. Transmural dispersion of repolarization in failing and nonfailing human ventricle. Circ Res. 2010;106:981-991. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 223] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 16. | Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol. 2012;9:208-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 267] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 17. | Suemizu H, Monnai M, Ohnishi Y, Ito M, Tamaoki N, Nakamura M. Identification of a key molecular regulator of liver metastasis in human pancreatic carcinoma using a novel quantitative model of metastasis in NOD/SCID/gammacnull (NOG) mice. Int J Oncol. 2007;31:741-751. [PubMed] [Cited in This Article: ] |
| 18. | Zhang L, Meissner E, Chen J, Su L. Current humanized mouse models for studying human immunology and HIV-1 immuno-pathogenesis. Sci China Life Sci. 2010;53:195-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927-933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 693] [Cited by in F6Publishing: 665] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 20. | Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D'Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, Tallquist MD. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118:400-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 757] [Cited by in F6Publishing: 869] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 21. | Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10:467-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 228] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 22. | Woodcock EA, Matkovich SJ. Cardiomyocytes structure, function and associated pathologies. Int J Biochem Cell Biol. 2005;37:1746-1751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | Zhou P, Pu WT. Recounting Cardiac Cellular Composition. Circ Res. 2016;118:368-370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 24. | Doppler SA, Carvalho C, Lahm H, Deutsch MA, Dreßen M, Puluca N, Lange R, Krane M. Cardiac fibroblasts: more than mechanical support. J Thorac Dis. 2017;9:S36-S51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 25. | Welser J. Primary Cells Versus Cell Lines [Internet]. ScienCell Research Laboratories. 2015;[cited 2018 Jun 30] Available from: https://sciencellonline.com/blog/primary-cells-versus-cell-lines/. [Cited in This Article: ] |
| 26. | Maqsood MI, Matin MM, Bahrami AR, Ghasroldasht MM. Immortality of cell lines: challenges and advantages of establishment. Cell Biol Int. 2013;37:1038-1045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 27. | Shaw SY, Brettman AD. Phenotyping patient-derived cells for translational studies in cardiovascular disease. Circulation. 2011;124:2444-2455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 28. | Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu Rev Genomics Hum Genet. 2011;12:217-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 29. | Vogenberg FR, Isaacson Barash C, Pursel M. Personalized medicine: part 1: evolution and development into theranostics. P T. 2010;35:560-576. [PubMed] [Cited in This Article: ] |
| 30. | Shay JW, Wright WE, Werbin H. Defining the molecular mechanisms of human cell immortalization. Biochim Biophys Acta. 1991;1072:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 116] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 31. | Yeh HH, Wu CH, Giri R, Kato K, Kohno K, Izumi H, Chou CY, Su WC, Liu HS. Oncogenic Ras-induced morphologic change is through MEK/ERK signaling pathway to downregulate Stat3 at a posttranslational level in NIH3T3 cells. Neoplasia. 2008;10:52-60. [PubMed] [Cited in This Article: ] |
| 32. | Hurlin PJ, Maher VM, McCormick JJ. Malignant transformation of human fibroblasts caused by expression of a transfected T24 HRAS oncogene. Proc Natl Acad Sci U S A. 1989;86:187-191. [PubMed] [Cited in This Article: ] |
| 33. | Kelekar A, Cole MD. Immortalization by c-myc, H-ras, and Ela oncogenes induces differential cellular gene expression and growth factor responses. Mol Cell Biol. 1987;7:3899-3907. [PubMed] [Cited in This Article: ] |
| 34. | Shay JW, Wright WE. Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp Cell Res. 1989;184:109-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 198] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3353] [Cited by in F6Publishing: 3543] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 36. | Metz T, Harris AW, Adams JM. Absence of p53 allows direct immortalization of hematopoietic cells by the myc and raf oncogenes. Cell. 1995;82:29-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 91] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 37. | Yamamoto A, Kumakura S, Uchida M, Barrett JC, Tsutsui T. Immortalization of normal human embryonic fibroblasts by introduction of either the human papillomavirus type 16 E6 or E7 gene alone. Int J Cancer. 2003;106:301-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Lee KM, Choi KH, Ouellette MM. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology. 2004;45:33-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 39. | Kavsan VM, Iershov AV, Balynska OV. Immortalized cells and one oncogene in malignant transformation: old insights on new explanation. BMC Cell Biol. 2011;12:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 40. | DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 402] [Cited by in F6Publishing: 375] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 41. | Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A-->G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet. 2001;68:238-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 162] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 42. | Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931-937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1079] [Cited by in F6Publishing: 1000] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 43. | Karicheva OZ, Kolesnikova OA, Schirtz T, Vysokikh MY, Mager-Heckel AM, Lombès A, Boucheham A, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011;39:8173-8186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 44. | Chuang YC, Liou CW, Chen SD, Wang PW, Chuang JH, Tiao MM, Hsu TY, Lin HY, Lin TK. Mitochondrial Transfer from Wharton's Jelly Mesenchymal Stem Cell to MERRF Cybrid Reduces Oxidative Stress and Improves Mitochondrial Bioenergetics. Oxid Med Cell Longev. 2017;2017:5691215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 45. | Wilkins HM, Carl SM, Swerdlow RH. Cytoplasmic hybrid (cybrid) cell lines as a practical model for mitochondriopathies. Redox Biol. 2014;2:619-631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 46. | Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van 't Veer-Korthof ET, Van der Harten JJ, Sobotka-Plojhar MA. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci. 1983;62:327-355. [PubMed] [Cited in This Article: ] |
| 47. | Ichida F, Tsubata S, Bowles KR, Haneda N, Uese K, Miyawaki T, Dreyer WJ, Messina J, Li H, Bowles NE, Towbin JA. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103:1256-1263. [PubMed] [Cited in This Article: ] |
| 48. | Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12:385-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 581] [Cited by in F6Publishing: 512] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 49. | Jefferies JL. Barth syndrome. Am J Med Genet C Semin Med Genet. 2013;163C:198-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 50. | Koshkin V, Greenberg ML. Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem J. 2002;364:317-322. [PubMed] [Cited in This Article: ] |
| 51. | Gonzalvez F, Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877-885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 203] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 52. | Bolhuis PA, Hensels GW, Hulsebos TJ, Baas F, Barth PG. Mapping of the locus for X-linked cardioskeletal myopathy with neutropenia and abnormal mitochondria (Barth syndrome) to Xq28. Am J Hum Genet. 1991;48:481-485. [PubMed] [Cited in This Article: ] |
| 53. | Gonzalez IL. Barth syndrome: TAZ gene mutations, mRNAs, and evolution. Am J Med Genet A. 2005;134:409-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 54. | Mullen PJ, Lüscher B, Scharnagl H, Krähenbühl S, Brecht K. Effect of simvastatin on cholesterol metabolism in C2C12 myotubes and HepG2 cells, and consequences for statin-induced myopathy. Biochem Pharmacol. 2010;79:1200-1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Burattini S, Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem. 2004;48:223-233. [PubMed] [Cited in This Article: ] |
| 56. | Lou W, Reynolds CA, Li Y, Liu J, Hüttemann M, Schlame M, Stevenson D, Strathdee D, Greenberg ML. Loss of tafazzin results in decreased myoblast differentiation in C2C12 cells: A myoblast model of Barth syndrome and cardiolipin deficiency. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:857-865. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 57. | Kaur G, Dufour JM. Cell lines: Valuable tools or useless artifacts. Spermatogenesis. 2012;2:1-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 394] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 58. | Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 59. | Ravikanth M, Soujanya P, Manjunath K, Saraswathi TR, Ramachandran CR. Heterogenecity of fibroblasts. J Oral Maxillofac Pathol. 2011;15:247-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 60. | Bainbridge P. Wound healing and the role of fibroblasts. J Wound Care. 2013;22:407-408, 410-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 370] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 61. | Alberts B, Johnson A, Lewis J, Raff M, Keith R, Walter P. Fibroblasts and Their Transformations: The Connective-Tissue Cell Family. In: Molecular Biology of the Cell. Garland Science; 2014; 1228-1232. [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 62. | Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 425] [Cited by in F6Publishing: 419] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 63. | Iwamiya T, Matsuura K, Masuda S, Shimizu T, Okano T. Cardiac fibroblast-derived VCAM-1 enhances cardiomyocyte proliferation for fabrication of bioengineered cardiac tissue. Regen Ther. 2016;4:92-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 64. | Baum J, Duffy HS. Fibroblasts and myofibroblasts: what are we talking about? J Cardiovasc Pharmacol. 2011;57:376-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 322] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 65. | Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 668] [Cited by in F6Publishing: 655] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 66. | Vangipuram M, Ting D, Kim S, Diaz R, Schüle B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. J Vis Exp. 2013;e3779. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 67. | Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4095] [Cited by in F6Publishing: 3887] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 68. | James AM, Wei YH, Pang CY, Murphy MP. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem J. 1996;318:401-407. [PubMed] [Cited in This Article: ] |
| 69. | Mancuso M. Antimyoclonic Effect of Levetiracetam in MERRF Syndrome. J Neurol Sci. 2006;243. [Cited in This Article: ] |
| 70. | Finsterer J. Central nervous system manifestations of mitochondrial disorders. Acta Neurol Scand. 2006;114:217-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 120] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 71. | D'Adamo P, Fassone L, Gedeon A, Janssen EA, Bione S, Bolhuis PA, Barth PG, Wilson M, Haan E, Orstavik KH, Patton MA, Green AJ, Zammarchi E, Donati MA, Toniolo D. The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet. 1997;61:862-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 190] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 72. | Schlame M, Kelley RI, Feigenbaum A, Towbin JA, Heerdt PM, Schieble T, Wanders RJ, DiMauro S, Blanck TJ. Phospholipid abnormalities in children with Barth syndrome. J Am Coll Cardiol. 2003;42:1994-1999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 145] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 73. | Fujioka H, Tandler B, Consolo MC, Karnik P. Division of mitochondria in cultured human fibroblasts. Microsc Res Tech. 2013;76:1213-1216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 74. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17055] [Article Influence: 947.5] [Reference Citation Analysis (0)] |
| 75. | Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634-7638. [PubMed] [Cited in This Article: ] |
| 76. | Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5956] [Cited by in F6Publishing: 5274] [Article Influence: 122.7] [Reference Citation Analysis (0)] |
| 77. | Raab S, Klingenstein M, Liebau S, Linta L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014;2014:768391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 78. | Singh VK, Kalsan M, Kumar N, Saini A, Chandra R. Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery. Front Cell Dev Biol. 2015;3:2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 236] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 79. | Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795-797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1249] [Cited by in F6Publishing: 1170] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 80. | Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 642] [Cited by in F6Publishing: 555] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 81. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1459] [Cited by in F6Publishing: 1301] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 82. | Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G, Kotton DN. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728-1740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 314] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 83. | Chen IP, Fukuda K, Fusaki N, Iida A, Hasegawa M, Lichtler A, Reichenberger EJ. Induced pluripotent stem cell reprogramming by integration-free Sendai virus vectors from peripheral blood of patients with craniometaphyseal dysplasia. Cell Reprogram. 2013;15:503-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 84. | Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1392] [Cited by in F6Publishing: 1231] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 85. | Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1350] [Cited by in F6Publishing: 1202] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 86. | Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1927] [Cited by in F6Publishing: 1831] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 87. | Rao MS, Malik N. Assessing iPSC reprogramming methods for their suitability in translational medicine. J Cell Biochem. 2012;113:3061-3068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 88. | Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, Cherry AB, Demirbas D, Tsankov AM, Zon LI, Rubin LL, Feinberg AP, Meissner A, Cowan CA, Daley GQ. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33:58-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 346] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 89. | Lai MI, Wendy-Yeo WY, Ramasamy R, Nordin N, Rosli R, Veerakumarasivam A, Abdullah S. Advancements in reprogramming strategies for the generation of induced pluripotent stem cells. J Assist Reprod Genet. 2011;28:291-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 90. | He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 402] [Cited by in F6Publishing: 381] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 91. | Shiraki N, Higuchi Y, Harada S, Umeda K, Isagawa T, Aburatani H, Kume K, Kume S. Differentiation and characterization of embryonic stem cells into three germ layers. Biochem Biophys Res Commun. 2009;381:694-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 92. | Yang C, Al-Aama J, Stojkovic M, Keavney B, Trafford A, Lako M, Armstrong L. Concise Review: Cardiac Disease Modeling Using Induced Pluripotent Stem Cells. Stem Cells. 2015;33:2643-2651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 93. | Chou SJ, Tseng WL, Chen CT, Lai YF, Chien CS, Chang YL, Lee HC, Wei YH, Chiou SH. Impaired ROS Scavenging System in Human Induced Pluripotent Stem Cells Generated from Patients with MERRF Syndrome. Sci Rep. 2016;6:23661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 94. | Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1672] [Cited by in F6Publishing: 1550] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 95. | Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524-528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1109] [Cited by in F6Publishing: 1044] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 96. | Tran TH, Wang X, Browne C, Zhang Y, Schinke M, Izumo S, Burcin M. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869-1878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 97. | Burridge PW, Diecke S, Matsa E, Sharma A, Wu H, Wu JC. Modeling Cardiovascular Diseases with Patient-Specific Human Pluripotent Stem Cell-Derived Cardiomyocytes. Methods Mol Biol. 2016;1353:119-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 98. | Lee JH, Protze SI, Laksman Z, Backx PH, Keller GM. Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell. 2017;21:179-194.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 236] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 99. | Rust W, Balakrishnan T, Zweigerdt R. Cardiomyocyte enrichment from human embryonic stem cell cultures by selection of ALCAM surface expression. Regen Med. 2009;4:225-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 100. | Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther. 2007;15:2027-2036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 101. | Yechikov S, Copaciu R, Gluck JM, Deng W, Chiamvimonvat N, Chan JW, Lieu DK. Same-Single-Cell Analysis of Pacemaker-Specific Markers in Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Subtypes Classified by Electrophysiology. Stem Cells. 2016;34:2670-2680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 102. | Rajala K, Pekkanen-Mattila M, Aalto-Setälä K. Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011;2011:383709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 103. | Machiraju P, Greenway SC. Current methods for the maturation of induced pluripotent stem cell-derived cardiomyocytes. World J Stem Cells. 2019;11:33-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 104. | Sun X, Nunes SS. Maturation of Human Stem Cell-derived Cardiomyocytes in Biowires Using Electrical Stimulation. J Vis Exp. 2017;123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 105. | Ruan JL, Tulloch NL, Razumova MV, Saiget M, Muskheli V, Pabon L, Reinecke H, Regnier M, Murry CE. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation. 2016;134:1557-1567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 283] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 106. | Correia C, Koshkin A, Duarte P, Hu D, Teixeira A, Domian I, Serra M, Alves PM. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci Rep. 2017;7:8590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 107. | Herron TJ, Rocha AM, Campbell KF, Ponce-Balbuena D, Willis BC, Guerrero-Serna G, Liu Q, Klos M, Musa H, Zarzoso M, Bizy A, Furness J, Anumonwo J, Mironov S, Jalife J. Extracellular Matrix-Mediated Maturation of Human Pluripotent Stem Cell-Derived Cardiac Monolayer Structure and Electrophysiological Function. Circ Arrhythm Electrophysiol. 2016;9:e003638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 108. | Moretti A, Laugwitz KL, Dorn T, Sinnecker D, Mummery C. Pluripotent stem cell models of human heart disease. Cold Spring Harb Perspect Med. 2013;3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 109. | Peter AK, Bjerke MA, Leinwand LA. Biology of the cardiac myocyte in heart disease. Mol Biol Cell. 2016;27:2149-2160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 110. | Kodaira M, Hatakeyama H, Yuasa S, Seki T, Egashira T, Tohyama S, Kuroda Y, Tanaka A, Okata S, Hashimoto H, Kusumoto D, Kunitomi A, Takei M, Kashimura S, Suzuki T, Yozu G, Shimojima M, Motoda C, Hayashiji N, Saito Y, Goto Y, Fukuda K. Impaired respiratory function in MELAS-induced pluripotent stem cells with high heteroplasmy levels. FEBS Open Bio. 2015;5:219-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 111. | Gropman AL. Diagnosis and treatment of childhood mitochondrial diseases. Curr Neurol Neurosci Rep. 2001;1:185-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 112. | Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848-E1857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1099] [Cited by in F6Publishing: 1151] [Article Influence: 95.9] [Reference Citation Analysis (0)] |
| 113. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13579] [Article Influence: 848.7] [Reference Citation Analysis (0)] |
| 114. | Warren L, Ni Y, Wang J, Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci Rep. 2012;2:657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 115. | Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, Van Biber B, Dardas T, Mignone JL, Izawa A, Hanna R, Viswanathan M, Gold JD, Kotlikoff MI, Sarvazyan N, Kay MW, Murry CE, Laflamme MA. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322-325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 562] [Cited by in F6Publishing: 521] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 116. | Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang DZ, Li K, Wang J, Wanders RJ, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616-623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 612] [Cited by in F6Publishing: 610] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 117. | Pandolfo M. Molecular basis of Friedreich ataxia. Mov Disord. 2001;16:815-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 118. | Weidemann F, Störk S, Liu D, Hu K, Herrmann S, Ertl G, Niemann M. Cardiomyopathy of Friedreich ataxia. J Neurochem. 2013;126 Suppl 1:88-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 119. | Minami I, Yamada K, Otsuji TG, Yamamoto T, Shen Y, Otsuka S, Kadota S, Morone N, Barve M, Asai Y, Tenkova-Heuser T, Heuser JE, Uesugi M, Aiba K, Nakatsuji N. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012;2:1448-1460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 120. | Crombie DE, Curl CL, Raaijmakers AJ, Sivakumaran P, Kulkarni T, Wong RC, Minami I, Evans-Galea MV, Lim SY, Delbridge L, Corben LA, Dottori M, Nakatsuji N, Trounce IA, Hewitt AW, Delatycki MB, Pera MF, Pébay A. Friedreich's ataxia induced pluripotent stem cell-derived cardiomyocytes display electrophysiological abnormalities and calcium handling deficiency. Aging (Albany NY). 2017;9:1440-1452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 121. | Greenway SC, Dallaire F, Hazari H, Patel D, Khan A. Addition of Digoxin Improves Cardiac Function in Children With the Dilated Cardiomyopathy With Ataxia Syndrome: A Mitochondrial Cardiomyopathy. Can J Cardiol. 2018;34:972-977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 122. | Ojala T, Polinati P, Manninen T, Hiippala A, Rajantie J, Karikoski R, Suomalainen A, Tyni T. New mutation of mitochondrial DNAJC19 causing dilated and noncompaction cardiomyopathy, anemia, ataxia, and male genital anomalies. Pediatr Res. 2012;72:432-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 123. | Rohani L, Meng G, Machiraju P, Liu S, Wu J, Kovalchuk I, Lewis I, Shutt TE, Khan A, Rancourt DE, Greenway S. Modeling the dilated cardiomyopathy with ataxia syndrome (DCMA), a pediatric mitochondrial cardiomyopathy, using cardiomyocytes derived from induced pluripotent stem cells. Can J Cardiol. 2017;33:S163–S164. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 124. | Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308-1320. [PubMed] [Cited in This Article: ] |
| 125. | Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 468] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 126. | Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GC, Tobita K, Rasmusson RL, Yang L. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res. 2014;104:258-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 136] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 127. | Pellman J, Zhang J, Sheikh F. Myocyte-fibroblast communication in cardiac fibrosis and arrhythmias: Mechanisms and model systems. J Mol Cell Cardiol. 2016;94:22-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 128. | Xie F, Gong K, Li K, Zhang M, Chang JC, Jiang S, Ye L, Wang J, Tan Y, Kan YW. Reversible Immortalization Enables Seamless Transdifferentiation of Primary Fibroblasts into Other Lineage Cells. Stem Cells Dev. 2016;25:1243-1248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 129. | Zhang Y, Nuglozeh E, Touré F, Schmidt AM, Vunjak-Novakovic G. Controllable expansion of primary cardiomyocytes by reversible immortalization. Hum Gene Ther. 2009;20:1687-1696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Davey KM, Parboosingh JS, McLeod DR, Chan A, Casey R, Ferreira P, Snyder FF, Bridge PJ, Bernier FP. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43:385-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 188] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 131. | Sparkes R, Patton D, Bernier F. Cardiac features of a novel autosomal recessive dilated cardiomyopathic syndrome due to defective importation of mitochondrial protein. Cardiol Young. 2007;17:215-217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 132. | Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2175] [Cited by in F6Publishing: 2085] [Article Influence: 208.5] [Reference Citation Analysis (0)] |
| 133. | Kim HS, Bernitz JM, Lee DF, Lemischka IR. Genomic editing tools to model human diseases with isogenic pluripotent stem cells. Stem Cells Dev. 2014;23:2673-2686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 134. | Marthaler AG, Schmid B, Tubsuwan A, Poulsen UB, Engelbrecht AF, Mau-Holzmann UA, Hyttel P, Nielsen JE, Nielsen TT, Holst B. Generation of an isogenic, gene-corrected control cell line of the spinocerebellar ataxia type 2 patient-derived iPSC line H271. Stem Cell Res. 2016;16:180-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 135. | Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 990] [Cited by in F6Publishing: 1082] [Article Influence: 90.2] [Reference Citation Analysis (0)] |