Abstract
Context:
Hepatitis C virus (HCV) infection is a major global public health issue. The Eastern Mediterranean regional office (EMRO) of the world health organization (WHO) seems to have one of the highest prevalence rates worldwide, with at least 21.3 million HCV-infected patients.Objectives:
The aim of the present study was to review systematically all epidemiological data related to the prevalence of HCV genotypes in infected patients in EMRO countries.Data Sources:
A systematic search was conducted of peer-reviewed journals indexed in electronic databases (PubMed, Scopus, ISI, PakMediNet, and IMEMR, and Persian-specific databases including SID, Iran Medex, and MagIran).Study Selection:
A systematic search was performed with temporal limits (papers published between January 2000 up to June 2015), regarding the prevalence and distribution of HCV genotypes in EMRO countries.Data Extraction:
The prevalence rates of HCV genotypes were pooled by metan command in Stata 14. Statistical heterogeneity was explored using the I-square at the 5% significance level. Publication bias was assessed, graphically and statistically, by funnel plot and Begg and Egger tests.Results:
A total of 563 records were identified through the electronic search. Of these records, 134 studies comprising 67681 HCV-infected individuals were included in the meta-analysis. In Iran, subtype 1a was the predominant subtype with a rate of 42% (95% CI, 39 - 46), followed by subtype 3a, 35% (95% CI, 31 - 38). In Pakistan, Subtype 3a was the most common subtype with a rate of 56% (95% CI, 49 - 62), followed by subtype 3b, 10% (95% CI, 7 - 12). In Saudi Arabia and Egypt, genotype 4 was the most prevalent genotype with a rate of 65% (95% CI, 59 - 72) and 69% (95% CI, 36 - 100) respectively. In Tunisia and Morocco, subtype 1b was the most common subtype with a rate of 69% (95% CI, 50 - 88) and 32% (95% CI, 7 - 56) respectively.Conclusions:
The genotype distribution of HCV takes diverse patterns in EMRO countries. Genotypes 1 and 3 were predominant in Iran and Pakistan, while genotype 4 and 1 were the most common genotypes in the Middle East Arab countries and North African Arab countries. Understanding the genotypes of HCV can help policy makers in designing good strategies for treatment.Keywords
Hepatitis C Virus Genotype Prevalence Molecular Epidemiology Systematic Review Meta-Analysis
1. Context
Hepatitis C virus (HCV) infection is a major global public health issue (1, 2). It is estimated that about 3% of the world’s population remains chronically infected with HCV, equaling almost 170 million individuals (3). Long-term chronic HCV infection can eventually lead to end-stage liver disease, cirrhosis, and hepatocellular carcinoma (4). Countries in the eastern Mediterranean regional office (EMRO) of the world health organization (WHO) seem to have one of the highest prevalence rates worldwide, with at least 21.3 million HCV-infected individuals (5).
Genetic variability is a distinctive feature of HCV, and viral sequences are currently classified into seven different genotypes and more than 67 subtypes (6). Viral genotypes may differ from each other in terms of response to standard antiviral therapy and geographical distribution (7, 8). Response rates to the standard treatment protocol of pegIFNα-2a and ribavirin are higher for patients infected with HCV genotypes 2 and 3 than for those with genotype 1 (8). The standard treatment regimen for chronic HCV infection can cost in excess of 22000 USD (9). Considering the high cost of treatment and the limited financial resources of many of the EMRO countries, more information about HCV genotype distribution in the EMRO can help policy makers make informed decisions about better resource allocation and programming priorities.
2. Objectives
The aim of the present study was to review systematically all epidemiological data related to the prevalence of HCV genotypes in infected patients in EMRO countries. A better knowledge about HCV genotype distribution in this region can promote better treatment strategies, and may improve the management of patients with HCV infection.
3. Data Sources
A systematic search was conducted of peer-reviewed journals indexed in PubMed, Scopus and ISI databases, reporting the prevalence and distribution of HCV genotypes in EMRO countries. In addition to the aforementioned databases, Persian-specific databases, including SID, Iran Medex and MagIran, the database of biomedical publications in Pakistan (PakMediNet), and the Index Medicus for the eastern Mediterranean region database (IMEMR) were also searched. The literature search was done with temporal limits (papers published between January 2000 up to June 2015) by using the following key words: hepatitis C virus or HCV, genotypes, genotype distribution, prevalence, and epidemiology. The names of EMRO countries that were added to our search strategy are as follows: Afghanistan, Bahrain, Djibouti, Egypt, Iran, Iraq, Jordan, Kuwait, Lebanon, Libya, Morocco, Oman, Pakistan, Palestine, Qatar, Saudi Arabia, Somalia, Sudan, Syria, Tunisia, United Arab Emirates, and Yemen.
4. Study Selection
All published studies fulfilling the following criteria were included in our analysis:1, studies in which the sample population was enrolled from EMRO countries with temporal limits (papers published between January 2000 up to June 2015); 2, full-text articles written in the English or Persian language on the HCV genotype prevalence among patients enrolled from EMRO countries; and 3, studies in the HCV-RNA-positive patients that used viral RNA for genotyping and relied on standard genotyping methods. The exclusion criteria were as follows: 1, studies with possible errors and unclear data; 2, studies in which only serologic findings were reported; 3, studies with non-primary data; and 4, any contraries to the inclusion criteria. All relevant papers obtained through the search strategy were reviewed by two independent reviewers (F.S) (M.S-V) and non-eligible studies were excluded (Kappa coefficient: 87.91). If there was any uncertainty, it was settled by consulting the supervisor of this investigation (S.M.A). Blinding and separation of tasks were considered in selection of studies and data extraction. The following indicators were extracted from each eligible study: first author’s name, year of publication, country of sample’s origin, sample size, risk factors, gender and mean age of subjects, genotype detection method, and HCV genotypes and subtypes.
5. Data Extraction
The prevalence rates of HCV genotypes were pooled by the metan command in Stata software version 14 (Stata Corp, College Station, TX, USA), and forest plot was used to display graphically the effect size of all the studies, with their confidence intervals, and the results of the meta-analysis. Statistical heterogeneity was explored using the I-square at the 5% significance level. The random effect model was used for pooling the studies’ prevalence rates. Publication bias was assessed graphically and statistically by funnel plot and Begg and Egger tests (metabias and metafunnel command in Stata) (10, 11). Funnel plots are a visual tool for investigating publication bias. They are simple scatter plots of the effect size estimated from individual studies (horizontal axis) against a measure of study size (vertical axis) in which standard error is used as the measure of study size. Publication bias may lead to asymmetrical funnel plots.
6. Results
6.1. Search Results and Study Selection
A total of 563 records were identified through the electronic search that reported HCV genotypes and subtypes from EMRO countries. Of these records, 349 duplicates were found and excluded. Based on screening of the remaining titles and abstracts, 158 potentially relevant manuscripts were retrieved and included for full full-text examination. We only included studies with primary data in our analysis; publications reporting the same data were considered duplicates and excluded. In addition, we excluded papers with unclear and ambiguous data. Screening of full-text studies identified 134 eligible studies for inclusion in the analysis (Figure 1).
Flowchart of Systematic Literature Search and Article Selection
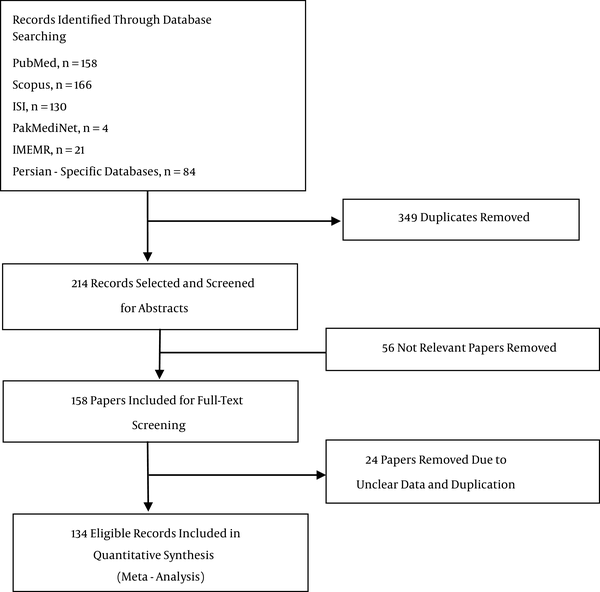
According to what we identified, no data were available from Djibouti, Oman, Somalia, and Yemen concerning prevalence of HCV genotypes and subtypes. Furthermore, only 1 study was found from Afghanistan, Palestine, Qatar, Syria, and Sudan. Finally, we found 49 studies from Iran involving 26058 subjects, 34 studies including 50407 subjects from Pakistan, 10 studies with 5270 subjects from Egypt, 6 studies with 2116 subjects from Saudi Arabia, 6 studies with 11825 subjects from Tunisia, 5 studies with 594 subjects from Iraq, 5 studies with 1680 subjects from Morocco, 3 studies with 374 subjects from Lebanon, 3 studies with 14641 subjects from Jordan, 2 studies with 264 subjects from Bahrain, 2 studies with 185 subjects from Kuwait, 2 studies with 2131 subjects from Libya, and 2 studies including 191 subjects from the United Arab Emirates. Populations investigated to estimate the prevalence of HCV genotypes and subtypes were mainly chronic-HCV-infected patients, such as people with thalassemia, hemophiliacs, hemodialysis patients, and injecting drug users. In addition, a few studies included general population groups, such as blood donors, healthy volunteers, and pregnant women. Characteristics of the included patients and the studies from the EMRO countries were shown in Appendix 1.
6.2. Prevalence of HCV Genotypes and Subtypes in EMRO Countries
Based on our results, genotype distribution of HCV takes diverse patterns in EMRO countries (Figure 2). According to the random-effect model, the estimated pooled prevalence of HCV genotypes and subtypes in each of the EMRO countries is listed below.
Distribution of Hepatitis C Virus Genotypes and Subtypes in EMRO Countries
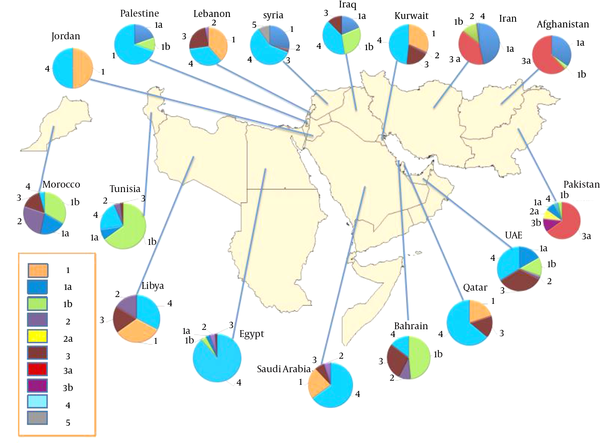
In Pakistan, genotype 3 was the predominant genotype with a rate of 67% (95% CI, 62 - 71); followed by genotype 1, 8% (95% CI, 6 - 10). Subtype 3a was the most common subtype with a rate of 56% (95% CI, 49 - 62); followed by subtype 3b, 10% (95% CI, 7 - 12). Appendix 2 provides forest plots depicting pooled summary estimates of the prevalence of HCV genotypes and subtypes in Pakistan. In Iran, genotype 1 was the most prevalent genotype with a rate of 54% (95% CI, 51 - 58); followed by genotype 3, 36% (95% CI, 32 - 39). Subtype 1a was the predominant subtype with a rate of 42% (95% CI, 39 - 46); followed by subtype 3a, 35% (95% CI, 31 - 38); and subtype 1b, 11% (95% CI, 10 - 13). Appendix 3 provides forest plots depicting pooled summary estimates of the prevalence of HCV genotypes and subtypes in Iran. A study in 113 HCV-RNA-positive injection drug users in Afghanistan showed subtype 3a as dominant with a rate of 39% (95% CI, 30 - 48); followed by subtype 1a, 22% (95% CI, 15 - 30).
Pooled summary estimates of the prevalence of HCV genotypes and subtypes in north African countries, including Egypt, Tunisia, Morocco, Libya, and Sudan, were shown in Appendix 4. In Tunisia, genotype 1 was the predominant genotype with a rate of 75% (95% CI, 57 - 94); followed by genotype 4, 22% (95% CI, 5 - 39). Subtype 1b was the most common subtype in Tunisia with a rate of 69% (95% CI, 50 - 88). In Egypt, genotype 4 was the most common genotype with a rate of 69% (95% CI, 36 - 100); followed by genotype 1, 5% (95% CI, 3 - 7). Subtype 4a was the most prevalent subtype with a rate of 59% (95% CI, 35 - 83); followed by subtype 1b, 3% (95% CI, 1 - 5). In Libya, genotype 4 and genotype 1 account for 32% (95% CI, 26 - 39); and 32% (95% CI, 30 - 34) of the pooled estimate of the HCV genotype rate, respectively, and were the predominant genotypes. There was no data about HCV subtypes in published studies from Libya. In Morocco, genotype 1 was the most prevalent genotype with a rate of 55% (95% CI, 41 - 69); followed by genotype 2, 26% (95% CI, 17 - 34). Subtype 1b was the most common subtype in Morocco with a rate of 32% (95% CI, 7 - 56). A study in 176 Sudanese patients with hepatosplenic schistosomiasis revealed genotype 4 as a major HCV genotype.
Pooled summary estimates of the prevalence of HCV genotypes and subtypes in Middle East Arab countries, including Saudi Arabia, Iraq, Lebanon, Jordan, Bahrain, Kuwait, the United Arab Emirates, Palestinian, Qatar, and Syria, were shown in Appendix 5. In Jordan, genotype 1 and genotype 4 account for 52% (95% CI, 11 - 94); and 52% (95% CI, 24 - 79) of the pooled estimate of the HCV genotype rate, respectively, and were the predominant genotypes. In Saudi Arabia, genotype 4 was the most prevalent genotype with a rate of 65% (95% CI, 59 - 72); followed by genotype 1, 23% (95% CI, 15 - 30). There was no data about HCV subtypes in published studies from Saudi Arabia. In Iraq, genotype 1 was the most common genotype with a rate of 45% (95% CI, 23 - 67); followed by genotype 4, 32% (95% CI, 17 - 48). Subtype 1b was the predominant subtype with a rate of 23% (95% CI, 12 - 35); followed by subtype 1a, 15% (95% CI, 2 - 27). In Lebanon, genotype 1 was the most common genotype with a rate of 37% (95% CI, 26 - 47); followed by genotype 4, 33% (95% CI, 18 - 47). The most common HCV genotype in Bahrain was genotype 1 with an estimated pooled prevalence of 37% (95% CI, 30 - 43). Genotype 4 was the most common genotype in Kuwait with a rate of 43% (95% CI, 33 - 52); followed by genotype 1, 28% (95% CI, 21 - 34). The predominant genotype in the United Arab Emirates was genotype 1 with a rate of 38% (95% CI, 16 - 60); followed by genotype 4, 30% (95% CI, 1 - 61). A study among 92 Palestinian chronic-HCV-infected patients reported genotype 4 as a predominant genotype with a rate of 64% (95% CI, 54 - 74); followed by genotype 1, 28% (95% CI, 19 - 37). An investigation into 400 chronic-HCV-infected patients in Qatar revealed genotype 4 as the most common genotype with a rate of 64% (95% CI, 59 - 69); followed by genotype 1, 20% (95% CI, 15 - 24). A comprehensive study among 636 consecutive HCV-infected patients referred to eight major medical centers in Syria showed genotype 4 as the most prevalent genotype with a rate of 59% (95% CI, 55 - 63); followed by genotype 1, 28% (95% CI, 25 - 32); and genotype 5, 10% (95% CI, 8 - 12).
7. Conclusions
The eastern Mediterranean regional office of the WHO comprises 22 countries, stretching from central Asia and the Middle East through north Africa, with a combined population of almost 583 million people. This region displays a heterogeneous distribution of HCV with at least 21.3 million HCV-infected patients (5). Determination of the most prevalent HCV genotypes in this region could be helpful to develop more effective treatment strategies against HCV infection. The present systematic review was conducted to shed more light on the distribution of HCV genotypes in EMRO countries. To determine HCV genotype prevalence in various countries, estimated pooled prevalence of HCV genotypes and subtypes were calculated for each of the EMRO countries. According to our results, genotype distribution of HCV takes diverse patterns in EMRO countries (Figure 2). In EMRO non-Arab countries (Iran, Pakistan and Afghanistan) genotypes 1 and 3 were predominant. In Iran, Subtype 1a was the most common subtype, followed by subtype 3a, which is similar to the HCV genotype distribution pattern in Northern Europe (12). In Pakistan subtype 3a was the most common subtype. There was only one report from Afghanistan, which showed predominance of subtype 3a, followed by subtype 1a (13). Due to war and political instability in Afghanistan, a tremendous number of Afghan immigrants usually make many trips between Iran, Afghanistan, and Pakistan (14), which may explain the HCV genotypes in Afghanistan. In the Middle East Arab countries genotypes 4 and 1 were predominant with two different distribution patterns; genotype 4 was the most common genotype in Saudi Arabia, Kuwait, Qatar, Syria, and Palestinian, while genotype 1 was found with higher frequency in Jordan, Lebanon, Iraq, Bahrain, and the United Arab Emirates. It should be noted that data from the Arabian Peninsula region has to be treated with caution due to the impact of foreign workers on HCV genotype prevalence. Furthermore, genotypes of HCV in North African Arab countries take two different distribution patterns; the first is limited to Nile river region (Egypt and Sudan) with genotype 4 being predominant, and the other pattern, which exists in the Maghreb region (Tunisia and Morocco), is characterized by the dominance of subtype 1b. Libya, in the Maghreb region, is the exception to this rule, as it shows a higher frequency of genotype 4 compared with Tunisia and Morocco. It is plausible that the Egyptian epidemic may have affected the HCV genotype distribution pattern in Libya, which is the neighboring country to the west of Egypt. Combination therapy with pegIFNα-2a and ribavirin is the most commonly available standard treatment protocol in EMRO countries, and the use of new direct-acting antiviral agents is not routine in most of the countries in this region. The clinical significance of HCV genotype distribution in EMRO countries is based on the impact of genotype on response to the standard treatment of pegIFNα-2a and ribavirin. The most common genotypes in EMRO countries, according to our results (genotypes 4 and 1) were also considered the most difficult to treat (8).
A proportion of HCV in EMRO countries’ patients was reported as untypeable. Untypeable genotypes represent HCV types other than those identified by the genotyping system, or a low viral gene copy number, which can escape genotype recognition. In order to recognize untypeable HCV genotypes, it is necessary to perform viral genome sequencing and phylogenetic analysis. In addition, mixed infections with different HCV genotypes can occur, and may lead to severe disease; low response rates to antiviral therapy increases the likelihood of relapse (15). Prevalence of infection with multiple HCV genotypes is variable between different countries in the EMRO region, and ranges from 0.3 in Iran to 0.13 in Iraq.
A number of limitations exist in the current study that should be noted. First, the sample size for some of the EMRO countries may not be adequate to produce a reliable estimate. Second, different HCV genotyping methods were utilized by various investigations, which may affect the obtained results of the present meta-analysis. Third, there is heterogeneity among the studies in terms of HCV-infected populations and patients’ risk groups, which makes the results more difficult to interpret. Furthermore, data deficiencies exist in some studies which did not allow for their analysis.
Taken together, the current study showed diverse patterns of HCV genotype distribution in EMRO countries. Genotypes 1 and 3 were predominant in Iran and Pakistan, while genotype 4 and 1 were the most common genotypes in the Middle East Arab countries and north African Arab countries. The most resistant types to treatment (genotypes 4 and 1) are found more commonly in EMRO countries, and greater effort is required to augment the treatment plans and strategies.
Supplements
Appendices 1 - 5 (13-145) are available at below link: http://hepatmon.com/?page=download&file_id=56809
Acknowledgements
References
-
1.
Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74-81. [PubMed ID: 19207969]. https://doi.org/10.1111/j.1478-3231.2008.01934.x.
-
2.
Alavian SM, Ahmadzad-Asl M, Lankarani KB, Shahbabaie MA, Bahrami AA, Kabir A. Hepatitis C infection in the general population of Iran: a systematic review. Hepat Mon. 2009;9(3):211-23.
-
3.
WHO. Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat. 1999;6(1):35-47. [PubMed ID: 10847128].
-
4.
Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349(9055):825-32. [PubMed ID: 9121257].
-
5.
WHO. Hepatitis C--global prevalence (update). Wkly Epidemiol Rec. 1999;74(49):425-7. [PubMed ID: 10645164].
-
6.
Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59(1):318-27. [PubMed ID: 24115039]. https://doi.org/10.1002/hep.26744.
-
7.
Bokharaei-Salim F, Keyvani H, Salehi-Vaziri M, Sadeghi F, Monavari SH, Mehrnoush L, et al. Mutations in the NS5A gene of hepatitis C virus subtype 1b and response to peg-IFNalpha-2a/RBV combination therapy in Azerbaijani patients. Arch Virol. 2014;159(11):2893-9. [PubMed ID: 25139545]. https://doi.org/10.1007/s00705-014-2133-0.
-
8.
Namazee N, Sali S, Asadi S, Shafiei M, Behnava B, Alavian SM. Real response to therapy in chronic hepatitis C virus patients: a study from iran. Hepat Mon. 2012;12(9). ee6151. [PubMed ID: 23087759]. https://doi.org/10.5812/hepatmon.6151.
-
9.
Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436(7053):946-52. [PubMed ID: 16107834]. https://doi.org/10.1038/nature04079.
-
10.
Harbord RM, Harris RJ, Sterne JAC. Updated tests for small-study effects in meta-analyses. Stata Journal. 2009;9(2):197.
-
11.
Sterne JAC, Harbord RM. Funnel plots in meta-analysis. Stata Journal. 2004;4:127-41.
-
12.
Khodabandehloo M, Roshani D. Prevalence of hepatitis C virus genotypes in Iranian patients: a systematic review and meta-analysis. Hepat Mon. 2014;14(12). ee22915. [PubMed ID: 25685164]. https://doi.org/10.5812/hepatmon.22915.
-
13.
Sanders-Buell E, Rutvisuttinunt W, Todd CS, Nasir A, Bradfield A, Lei E, et al. Hepatitis C genotype distribution and homology among geographically disparate injecting drug users in Afghanistan. J Med Virol. 2013;85(7):1170-9. [PubMed ID: 23918535]. https://doi.org/10.1002/jmv.23575.
-
14.
Rastin M, Mahmoudi M, Rezaee SA, Assarehzadegan MA, Tabasi N, Zamani S, et al. Distribution of Hepatitis C virus genotypes in city of Mashhad, North-east of Iran. Indian J Med Microbiol. 2014;32(1):53-6. [PubMed ID: 24399389]. https://doi.org/10.4103/0255-0857.124306.
-
15.
Zali MR, Mayumi M, Raoufi M, Nowroozi A. Hepatitis C virus genotypes in the Islamic Republic of Iran: a preliminary study. East Mediterr Health J. 2000;6(2-3):372-7. [PubMed ID: 11556026].
-
16.
Samimi-Rad K, Nategh R, Malekzadeh R, Norder H, Magnius L. Molecular epidemiology of hepatitis C virus in Iran as reflected by phylogenetic analysis of the NS5B region. J Med Virol. 2004;74(2):246-52. [PubMed ID: 15332273]. https://doi.org/10.1002/jmv.20170.
-
17.
Kazemi B, Tafvizi F, Bandehpour M. Determination of HCV genotypes in Iran. Biotechnology. 2005;4(2):139-43.
-
18.
Ahmadi Pour MH, Keivani H, Sabahi F, Alavian SM. Determination of HCV Genotypes, in Iran by PCR-RFLP. Iranian J Publ Health. 2006;35(4):54-61.
-
19.
Hosseini-Moghaddam SM, Keyvani H, Kasiri H, Kazemeyni SM, Basiri A, Aghel N, et al. Distribution of hepatitis C virus genotypes among hemodialysis patients in Tehran--a multicenter study. J Med Virol. 2006;78(5):569-73. [PubMed ID: 16555284]. https://doi.org/10.1002/jmv.20577.
-
20.
Kabir A, Alavian SM, Keyvani H. Distribution of hepatitis C virus genotypes in patients infected by different sources and its correlation with clinical and virological parameters: a preliminary study. Comp Hepatol. 2006;5(1):1.
-
21.
Hejazi MS, Ghotaslou R, Farshdoosty Hagh M, Mohammadzadeh Sadigh Y. Genotyping of Hepatitis C virus in northwest of Iran. Biotech. 2007;6(3):302-8. https://doi.org/10.3923/biotech.2007.302.308.
-
22.
Keyvani H, Alizadeh AH, Alavian SM, Ranjbar M, Hatami S. Distribution frequency of hepatitis C virus genotypes in 2231 patients in Iran. Hepatol Res. 2007;37(2):101-3. [PubMed ID: 17300704]. https://doi.org/10.1111/j.1872-034X.2007.00015.x.
-
23.
Samimi-Rad K, Shahbaz B. Hepatitis C virus genotypes among patients with thalassemia and inherited bleeding disorders in Markazi province, Iran. Haemophilia. 2007;13(2):156-63. [PubMed ID: 17286768]. https://doi.org/10.1111/j.1365-2516.2006.01415.x.
-
24.
Samimi-Rad K, Hosseini M, Shahbaz B. Hepatitis C virus infection and hcv genotypes of hemodialysis patients. IJPH. 2008;37(3):146-52.
-
25.
Sharifi Z, Mahmoudian Shooshtari M. Hepatitis C virus infection and genotypes in blood donors. Iran J Virol. 2008;2(4):17-22.
-
26.
Somi MH, Keivani H, Ardalan MR, Farhang S, Pouri AA. Hepatitis C virus genotypes in patients with end-stage renal disease in East Azerbaijan, Iran. Saudi J Kidney Dis Transpl. 2008;19(3):461-5. [PubMed ID: 18445914].
-
27.
Alavian SM, Miri SM, Keshvari M, Elizee PK, Behnava B, Tabatabaei SV, et al. Distribution of hepatitis C virus genotype in Iranian multiply transfused patients with thalassemia. Transfusion. 2009;49(10):2195-9. [PubMed ID: 19538541]. https://doi.org/10.1111/j.1537-2995.2009.02252.x.
-
28.
Amini S, MahmoodMahmoodiFarahani MA, Mahsa J, MalekHossein A. Distribution of hepatitis c virus genotypes in iran: A population-based study. Hepat mon. 2009;2009(2, Spring):95-102.
-
29.
Assarehzadegan MA, Shakerinejad G, Norouzirad R, Amini A. Distribution of hepatitis C virus genotypes among patients with hepatitis C infection in Khuzestan province. AJUMS. 2009;7(4):Pe471-8.
-
30.
Assarehzadegan MA, Shakerinejad G, Noroozkohnejad R, Amini A, Rahim Rezaee SA. Prevalence of hepatitis C and B infection and HC V genotypes among hemodialysis patients in Khuzestan province, southwest Iran. Saudi J Kidney Dis Transpl. 2009;20(4):681-4. [PubMed ID: 19587521].
-
31.
Davarpanah MA, Saberi-Firouzi M, Bagheri Lankarani K, Mehrabani D, Behzad Behbahani A, Serati A, et al. Hepatitis C virus genotype distribution in Shiraz, southern Iran. Hepat Mon. 2009;9(2):122-7.
-
32.
Bokharaei Salim F, Keyvani H, Amiri A, Jahanbakhsh Sefidi F, Shakeri R, Zamani F. Distribution of different hepatitis C virus genotypes in patients with hepatitis C virus infection. World J Gastroenterol. 2010;16(16):2005-9. [PubMed ID: 20419838].
-
33.
Farshadpour F, Makvandi M, Samarbafzadeh AR, Jalalifar MA. Determination of hepatitis C virus genotypes among blood donors in Ahvaz, Iran. Indian J Med Microbiol. 2010;28(1):54-6. [PubMed ID: 20061766]. https://doi.org/10.4103/0255-0857.58731.
-
34.
Hajia M, Amirzargar A, Khedmat H, Shahrokhi N, Farzanehkhah M, Ghorishi S, et al. Genotyping Pattern among Iranian HCV Positive Patients. Iran J Public Health. 2010;39(2):39-44. [PubMed ID: 23113005].
-
35.
Keshvari M, Alavian SM, Behnava B, Miri SM, Tabatabaei SV, Abolghasemi H, et al. Distribution of hepatitis C virus genotypes in iranian patients with congenital bleeding disorders. IRCMJ. 2010;2010(6):608-14.
-
36.
Shahraki T, Shahraki M, Moghaddam ES, Najafi M, Bahari A. Determination of hepatitis C genotypes and the viral titer distribution in children and adolescents with major thalassemia. Iran J Pediatr. 2010;20(1):75-81. [PubMed ID: 23056686].
-
37.
Sharifi Z, Shooshtari MM, Kermani FR. Identification of HCV genotypes in HCV infected blood donors. Indian J Microbiol. 2010;50(3):275-9. [PubMed ID: 23100841]. https://doi.org/10.1007/s12088-010-0059-0.
-
38.
Vahdat K, Keyvani H, Tabib SM, Rostamabadi S, Valizadeh SM, Cheraghi S, et al. Molecular epidemiology of hepatitis C virus genotypes in Bushehr province, Iran. Eur Rev Med Pharmacol Sci. 2010;14(10):861-4. [PubMed ID: 21222372].
-
39.
Zarkesh-Esfahani SH, Kardi MT, Edalati M. Hepatitis C virus genotype frequency in Isfahan province of Iran: a descriptive cross-sectional study. Virol J. 2010;7:69. [PubMed ID: 20331907]. https://doi.org/10.1186/1743-422X-7-69.
-
40.
Farivar TN, Nezam MK, Johari P. Genotyping of hepatitis C virus isolated from hepatitis patients in Southeast of Iran by Taqman Realtime PCR. J Pak Med Assoc. 2011;61(6):586-8. [PubMed ID: 22204216].
-
41.
Hajiani E, Hashemi SJ, Masjedizadeh A, Shayesteh AA, Jalali F. Genotypic analysis of hepatitis C virus in khuzestan province, southwestern iran. Middle East J Dig Dis. 2011;3(2):126-30. [PubMed ID: 25197544].
-
42.
Moradi A, Semnani S, Keshtkar A, Khodabakhshi B, Kazeminejad V, Molana A, et al. Distribution of hepatitis C virus genotype among HCV infected patients in Golestan province, Iran. Govaresh. 2011;15(1):7-13.
-
43.
Tajbakhsh E, Dosti A, Tajbakhsh S, Momeni M, Tajbakhsh F. Determination of hepatitis C virus genotypes among HCV positive patients in Shahrekord, Iran. Afr J Microbiol Res. 2011;5(32):5910-5.
-
44.
Ziyaeyan M, Abdolvahab A, Marziyeh J, Parisa B, Mahsa M, Ali K. Prevalence of hepatitis C virus genotypes in chronic infected patients, southern Iran. Jundishapur J Microbiol. 2016;2011(3, Summer).
-
45.
Assarehzadegan MA, Ghafourian Boroujerdnia M, Zandian K. Prevalence of hepatitis B and C infections and HCV genotypes among haemophilia patients in ahvaz, southwest iran. Iran Red Crescent Med J. 2012;14(8):470-4. [PubMed ID: 23105982].
-
46.
Ghane M, Eghbali M, Nejad HR, Saeb K, Farahani M. Distribution of hepatitis C virus genotypes amongst the beta-thalassemia patients in North of Iran. Pak J Biol Sci. 2012;15(15):748-53. [PubMed ID: 24171261].
-
47.
Haghshenas M, Babamahmoodi F, Rafiei A, Vahedi V, Alizadeh-Navaei R. Prevalence of Hepatitis C virus genotypes in the Northern of Iran (Mazandaran) from 2009 to 2011. Healthmed. 2012;6(9):3046-50.
-
48.
Joukar F, Khalesi AK, Jafarshad R, Rahimabadi MS, Mansour-Ghanaei F. Distribution of hepatitis C virus genotypes in haemodialysis patients of Guilan, northern Islamic Republic of Iran. East Mediterr Health J. 2012;18(3):236-40. [PubMed ID: 22574476].
-
49.
Karimi A, Imani R, Karti M, Taheri S. Frequency of hepatitis C virus genotypes in infected patients in Charmahal-O-Bakhtiari Province, Iran. Afr J Microbiol Res. 2012;6(9):2230-2.
-
50.
Vossughinia H, Goshayeshi L, Bayegi HR, Sima H, Kazemi A, Erfani S, et al. Prevalence of hepatitis C virus genotypes in mashhad, northeast iran. Iran J Public Health. 2012;41(9):56-61. [PubMed ID: 23193507].
-
51.
Jahanbakhsh Sefidi F, Keyvani H, Monavari SH, Alavian SM, Fakhim S, Bokharaei-Salim F. Distribution of hepatitis C virus genotypes in Iranian chronic infected patients. Hepat Mon. 2013;13(1). ee7991. [PubMed ID: 23550108]. https://doi.org/10.5812/hepatmon.7991.
-
52.
Kermani FR, Sharifi Z, Ferdowsian F, Paz Z, Zamanian M. Distribution of Hepatitis C Virus Genotypes Among Chronic Infected Injecting Drug Users in Tehran, Iran. Jundishapur J Microbiol. 2013;6(3):265-8.
-
53.
Mousavi SF, Moosavy SH, Alavian SM, Eghbali H, Mahboobi H. Distribution of hepatitis C virus genotypes among patients with hepatitis C virus infection in hormozgan, iran. Hepat Mon. 2013;13(12). ee14324. [PubMed ID: 24403914]. https://doi.org/10.5812/hepatmon.14324.
-
54.
Rafiei A, Darzyani AM, Taheri S, Haghshenas MR, Hosseinian A, Makhlough A. Genetic diversity of HCV among various high risk populations (IDAs, thalassemia, hemophilia, HD patients) in Iran. Asian Pac J Trop Med. 2013;6(7):556-60. [PubMed ID: 23768829]. https://doi.org/10.1016/S1995-7645(13)60096-6.
-
55.
Samimi-Rad K, Asgari F, Nasiritoosi M, Esteghamati A, Azarkeyvan A, Eslami SM, et al. Patient-to-Patient Transmission of Hepatitis C at Iranian Thalassemia Centers Shown by Genetic Characterization of Viral Strains. Hepat Mon. 2013;13(1). ee7699. [PubMed ID: 23585766]. https://doi.org/10.5812/hepatmon.7699.
-
56.
Afshari R, Nomani H, Zaniani FR, Nabavinia MS, Mirbagheri Z, Meshkat M, et al. Genotype distribution of hepatitis C virus in Khorasan Razavi Province, Iran. Turk J Med Sci. 2014;44(4):656-60. [PubMed ID: 25551938].
-
57.
Ashrafi Hafez A, Baharlou R, Mousavi Nasab SD, Ahmadi Vasmehjani A, Shayestehpour M, Joharinia N, et al. Molecular epidemiology of different hepatitis C genotypes in serum and peripheral blood mononuclear cells in jahrom city of iran. Hepat Mon. 2014;14(5). ee16391. [PubMed ID: 24976833]. https://doi.org/10.5812/hepatmon.16391.
-
58.
Esmaeilzadeh A, Erfanmanesh M, Ghasemi S, Mohammadi F. Serological assay and genotyping of hepatitis C virus in infected patients in zanjan province. Hepat Mon. 2014;14(9). ee17323. [PubMed ID: 25368655]. https://doi.org/10.5812/hepatmon.17323.
-
59.
Hadinedoushan H, Salmanroghani H, Amirbaigy MK, Akhondi-Meybodi M. Hepatitis C virus genotypes and association with viral load in yazd, central province of iran. Hepat Mon. 2014;14(3). ee11705. [PubMed ID: 24693314]. https://doi.org/10.5812/hepatmon.11705.
-
60.
Jamalidoust M, Namayandeh M, Asaei S, Aliabadi N, Ziyaeyan M. Determining hepatitis C virus genotype distribution among high-risk groups in Iran using real-time PCR. World J Gastroenterol. 2014;20(19):5897-902. [PubMed ID: 24914351]. https://doi.org/10.3748/wjg.v20.i19.5897.
-
61.
Salehi Moghadam F, Mohebbi SR, Hosseini SM, Romani S, Mirtalebi H, Azimzadeh P, et al. Phylogenetic analysis of hepatitis C virus strains and risk factors associated with infection and viral subtypes among Iranian patients. J Med Virol. 2014;86(8):1342-9. [PubMed ID: 24838700]. https://doi.org/10.1002/jmv.23947.
-
62.
Shanehsazzadeh M, Rad J, Pourazar A, Behbahani M. Frequency distribution of hepatitis c virus (hcv) genotypes and its association with viral loads in chronic hcv infected patients of isfahan, iran. J APPL MICROBIOL.
-
63.
Qazi MA, Fayyaz M, Chaudhary GMD, Jamil A, Malik A, Gardezi AI, et al. Hepatitis C virus genotypes in Bahawalpur. Biomedica. 2006;22:51-4.
-
64.
Ahmad N, Asgher M, Shafique M, Qureshi JA. An evidence of high prevalence of Hepatitis C virus in Faisalabad, Pakistan. Saudi Med J. 2007;28(3):390-5. [PubMed ID: 17334466].
-
65.
Hakim S, Kazmi S, Bagasra O. Seroprevalence of hepatitis B and C genotypes among young apparently healthy females of karachi-pakistan. Libyan J Med. 2008;3(2):66-70. [PubMed ID: 21499460]. https://doi.org/10.4176/071123.
-
66.
Idrees M, Riazuddin S. Frequency distribution of hepatitis C virus genotypes in different geographical regions of Pakistan and their possible routes of transmission. BMC Infect Dis. 2008;8:69. [PubMed ID: 18498666]. https://doi.org/10.1186/1471-2334-8-69.
-
67.
Mumtaz K, Hamid SS, Moatter T, Abid S, Shah HA, Jafri W. Distribution of hepatitis C virus genotypes and its response to treatment in Pakistani patients. Saudi Med J. 2008;29(11):1671-3. [PubMed ID: 18998024].
-
68.
Abbas SZ, Ali M, Muhammad AH, Shaw S, Abbas SQ. Frequency of HCV infection and its genotypes among patients attending a liver clinic and voluntary blood donors in a rural area of Pakistan. Pak J Med Sci. 2009;25(4):579-82.
-
69.
Afridi S, Naeem M, Hussain A, Kakar N, Babar ME, Ahmad J. Prevalence of hepatitis C virus (HCV) genotypes in Balochistan. Mol Biol Rep. 2009;36(6):1511-4. [PubMed ID: 18766467]. https://doi.org/10.1007/s11033-008-9342-0.
-
70.
Husain A, Malik FA, Nagra H, Ehsan A, Ahmad Z, Abid M. Frequency of different HCV genotypes in Faisalabad. APMC. 2009;3(1):19-22.
-
71.
Idrees M, Butt S, Awan Z, Aftab M, Khubaib B, Rehman IU, et al. Nucleotide identity and variability among different Pakistani hepatitis C virus isolates. Virol J. 2009;6:130. [PubMed ID: 19698187]. https://doi.org/10.1186/1743-422X-6-130.
-
72.
Idrees M, Rafique S, Rehman I, Akbar H, Yousaf MZ, Butt S, et al. Hepatitis C virus genotype 3a infection and hepatocellular carcinoma: Pakistan experience. World J Gastroenterol. 2009;15(40):5080-5. [PubMed ID: 19860002].
-
73.
Khan A, Tanaka Y, Azam Z, Abbas Z, Kurbanov F, Saleem U, et al. Epidemic spread of hepatitis C virus genotype 3a and relation to high incidence of hepatocellular carcinoma in Pakistan. J Med Virol. 2009;81(7):1189-97. [PubMed ID: 19475617]. https://doi.org/10.1002/jmv.21466.
-
74.
Yasmeen A, Siddiqui AA, Hamid S, Sultana T, Jafri W, Persson MA. Genetic variations in a well conserved 5'-untranslated region of hepatitis C virus genome isolated in Pakistan. J Virol Methods. 2009;160(1-2):38-47. [PubMed ID: 19406160]. https://doi.org/10.1016/j.jviromet.2009.04.007.
-
75.
Ahmad W, Ijaz B, Javed FT, Jahan S, Shahid I, Khan FM, et al. HCV genotype distribution and possible transmission risks in Lahore, Pakistan. World J Gastroenterol. 2010;16(34):4321-8. [PubMed ID: 20818816].
-
76.
Ali A, Ahmed H, Idrees M. Molecular epidemiology of Hepatitis C virus genotypes in Khyber Pakhtoonkhaw of Pakistan. Virol J. 2010;7:203. [PubMed ID: 20796270]. https://doi.org/10.1186/1743-422X-7-203.
-
77.
Butt S, Idrees M, Akbar H, ur Rehman I, Awan Z, Afzal S, et al. The changing epidemiology pattern and frequency distribution of hepatitis C virus in Pakistan. Infect Genet Evol. 2010;10(5):595-600. [PubMed ID: 20438863]. https://doi.org/10.1016/j.meegid.2010.04.012.
-
78.
Mahmood K, Mohammad N. Genotype variation of hepatitis C virus in District Buner Swat. JAMC. 2010;23(4):18-21.
-
79.
Safi S, Badshah Y, Waheed Y, Fatima K, Tahir S, Shinwari A, et al. Distribution of hepatitis C virus genotypes, hepatic steatosis and their correlation with clinical and virological factors in Pakistan. Asian Biomed. 2010;4(2).
-
80.
Ali A, Nisar M, Ahmad H, Saif N, Idrees M, Bajwa MA. Determination of HCV genotypes and viral loads in chronic HCV infected patients of Hazara Pakistan. Virol J. 2011;8:466. [PubMed ID: 21982599]. https://doi.org/10.1186/1743-422X-8-466.
-
81.
Ali S, Ali I, Azam S, Ahmad B. Frequency distribution of HCV genotypes among chronic hepatitis C patients of Khyber Pakhtunkhwa. Virol J. 2011;8:193. [PubMed ID: 21521506]. https://doi.org/10.1186/1743-422X-8-193.
-
82.
Ijaz B, Ahmad W, Javed FT, Gull S, Sarwar MT, Kausar H, et al. Association of laboratory parameters with viral factors in patients with hepatitis C. Virology journal. 2011;8(1):1.
-
83.
Inamullah, Idrees M, Ahmed H, Sajid UG, Ali M, Ali L, et al. Hepatitis C virus genotypes circulating in district Swat of Khyber Pakhtoonkhaw, Pakistan. Virol J. 2011;8:16. [PubMed ID: 21235746]. https://doi.org/10.1186/1743-422X-8-16.
-
84.
Khan S, Attaullah S, Ayaz S, Niaz Khan S, Shams S, Ali I, et al. Molecular epidemiology of hcv among health care workers of khyber pakhtunkhwa. Virol J. 2011;8:105. [PubMed ID: 21385397]. https://doi.org/10.1186/1743-422X-8-105.
-
85.
Nawaz M, Siddique S, Manzoor I, Nadeem S, Din S, Shabbir H, et al. Infection of hepatitis C virus genotypes in hepatocellular carcinoma patients from rural areas of Faisalabad region, Pakistan. Afr J Biotechnol. 2013;10(42):8471-5.
-
86.
Rehman L, Ullah I, Ali I, Khan IA, Iqbal A, Khan S, et al. Active hepatitis C infection and HCV genotypes prevalent among the IDUs of Khyber Pakhtunkhwa. Virol J. 2011;8:327. [PubMed ID: 21711541]. https://doi.org/10.1186/1743-422X-8-327.
-
87.
Safi AZ, Waheed Y, Sadat J, Salahuddin S, Saeed U, Ashraf M. Molecular study of HCV detection, genotypes and their routes of transmission in North West Frontier Province, Pakistan. Asian Pacific journal of tropical biomedicine. 2012;2(7):532-6.
-
88.
Afridi SQ, Zahid MN, Shabbir MZ, Hussain Z, Mukhtar N, Tipu MY, et al. Prevalence of HCV genotypes in district Mardan. Virol J. 2013;10:90. [PubMed ID: 23514695]. https://doi.org/10.1186/1743-422X-10-90.
-
89.
Aziz H, Raza A, Murtaza S, Waheed Y, Khalid A, Irfan J, et al. Molecular epidemiology of hepatitis C virus genotypes in different geographical regions of Punjab Province in Pakistan and a phylogenetic analysis. Int J Infect Dis. 2013;17(4):247-53. [PubMed ID: 23183233]. https://doi.org/10.1016/j.ijid.2012.09.017.
-
90.
Bashir MF, Haider MS, Rashid N, Riaz S. Core gene expression and association of genotypes with viral load in hepatitis C virus (HCV)-infected patients in Punjab, Pakistan. TROP J PHARM RES. 2013;12(3):335-41.
-
91.
Rauf A, Nadeem M, Arshad M, Riaz H, Latif M, Iqbal M, et al. Prevalence of Hepatitis B and C Virus in the General Population of Hill Surang Area, Azad Jammu and Kashmir, Pakistan. PAK J ZOOL. 2013;45(2):543-8.
-
92.
Afridi SQ, Ali MM, Awan F, Zahid MN, Afridi IQ, Afridi SQ, et al. Molecular epidemiology and viral load of HCV in different regions of Punjab, Pakistan. Virol J. 2014;11:24. [PubMed ID: 24512668]. https://doi.org/10.1186/1743-422X-11-24.
-
93.
Ali S, Ahmad A, Khan RS, Khan S, Hamayun M, Khan SA, et al. Genotyping of HCV RNA reveals that 3a is the most prevalent genotype in mardan, pakistan. Adv Virol. 2014;2014:606201. [PubMed ID: 24715902]. https://doi.org/10.1155/2014/606201.
-
94.
Khan N, Akmal M, Hayat M, Umar M, Ullah A, Ahmed I, et al. Geographic distribution of hepatitis C virus genotypes in pakistan. Hepat Mon. 2014;14(10). ee20299. [PubMed ID: 25477975]. https://doi.org/10.5812/hepatmon.20299.
-
95.
Saleha S, Kamal A, Ullah F, Khan N, Mahmood A, Khan S. Prevalence of hepatitis C virus genotypes in district bannu, khyber pakhtunkhwa, pakistan. Hepat Res Treat. 2014;2014:165826. [PubMed ID: 25093120]. https://doi.org/10.1155/2014/165826.
-
96.
Waqar M, Khan AU, Rehman HU, Idrees M, Wasim M, Ali A, et al. Determination of hepatitis C virus genotypes circulating in different districts of Punjab (Pakistan). Eur J Gastroenterol Hepatol. 2014;26(1):59-64. [PubMed ID: 23743560]. https://doi.org/10.1097/MEG.0b013e328362dc3f.
-
97.
Kamal S, Madwar M, Bianchi L, Tawil AE, Fawzy R, Peters T, et al. Clinical, virological and histopathological features: long-term follow-up in patients with chronic hepatitis C co-infected with S. mansoni. Liver. 2000;20(4):281-9. [PubMed ID: 10959806].
-
98.
Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout egypt. J Infect Dis. 2000;182(3):698-707. [PubMed ID: 10950762]. https://doi.org/10.1086/315786.
-
99.
Tanaka Y, Agha S, Saudy N, Kurbanov F, Orito E, Kato T, et al. Exponential spread of hepatitis C virus genotype 4a in Egypt. J Mol Evol. 2004;58(2):191-5. [PubMed ID: 15042339]. https://doi.org/10.1007/s00239-003-2541-3.
-
100.
Elsawy EM, Sobh MA, El-Chenawi FA, Hassan IM, Shehab El-Din AB, Ghoneim MA. Serotyping of hepatitis C virus in hemodialysis patients: comparison with a standardized genotyping assay. Diagn Microbiol Infect Dis. 2005;51(2):91-4. [PubMed ID: 15698713]. https://doi.org/10.1016/j.diagmicrobio.2004.09.010.
-
101.
Abdel-Hamid M, El-Daly M, Molnegren V, El-Kafrawy S, Abdel-Latif S, Esmat G, et al. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88(5):1526-31. [PubMed ID: 17412982]. https://doi.org/10.1099/vir.0.82626-0.
-
102.
Elkady A, Tanaka Y, Kurbanov F, Sugauchi F, Sugiyama M, Khan A, et al. Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication. J Med Virol. 2009;81(6):1015-23. [PubMed ID: 19382263]. https://doi.org/10.1002/jmv.21492.
-
103.
Omran MH, Youssef SS, El-Garf WT, Tabll AA, Bader-Eldin NG, Atef K, et al. Phylogenetic and genotyping of hepatitis C virus in Egypt. Aust J Basic Appl Sci. 2009;3(1):1-8.
-
104.
Ryu SH, Fan X, Xu Y, Elbaz T, Zekri AR, Abdelaziz AO, et al. Lack of association between genotypes and subtypes of HCV and occurrence of hepatocellular carcinoma in Egypt. J Med Virol. 2009;81(5):844-7. [PubMed ID: 19319951]. https://doi.org/10.1002/jmv.21451.
-
105.
Youssef A, Yano Y, Utsumi T, abd El-alah EM, abd El-Hameed Ael E, Serwah Ael H, et al. Molecular epidemiological study of hepatitis viruses in Ismailia, Egypt. Intervirology. 2009;52(3):123-31. [PubMed ID: 19468235]. https://doi.org/10.1159/000219385.
-
106.
Elsadek Fakhr A, Pourkarim MR, Maes P, Atta AH, Marei A, Azab M, et al. Hepatitis C Virus NS5B Sequence-Based Genotyping Analysis of Patients From the Sharkia Governorate, Egypt. Hepat Mon. 2013;13(12). ee12706. [PubMed ID: 24358038]. https://doi.org/10.5812/hepatmon.12706.
-
107.
Sassi F, Gorgi Y, Ayed K, Abdallah TB, Lamouchi A, Maiz HB. Hepatitis C virus antibodies in dialysis patients in Tunisia: a single center study. Saudi J Kidney Dis Transpl. 2000;11(2):218-22. [PubMed ID: 18209319].
-
108.
Djebbi A, Triki H, Bahri O, Cheikh I, Sadraoui A, Ben Ammar A, et al. Genotypes of hepatitis C virus circulating in Tunisia. Epidemiol Infect. 2003;130(3):501-5. [PubMed ID: 12825736].
-
109.
Othman SB, Trabelsi A, Monnet A, Bouzgarrou N, Grattard F, Beyou A, et al. Evaluation of a prototype HCV NS5b assay for typing strains of hepatitis C virus isolated from Tunisian haemodialysis patients. J Virol Methods. 2004;119(2):177-81. [PubMed ID: 15158600]. https://doi.org/10.1016/j.jviromet.2004.03.002.
-
110.
Bouzgarrou N, Fodha I, Othman SB, Achour A, Grattard F, Trabelsi A, et al. Evaluation of a total core antigen assay for the diagnosis of hepatitis C virus infection in hemodialysis patients. J Med Virol. 2005;77(4):502-8. [PubMed ID: 16254976]. https://doi.org/10.1002/jmv.20485.
-
111.
Mejri S, Salah AB, Triki H, Alaya NB, Djebbi A, Dellagi K. Contrasting patterns of hepatitis C virus infection in two regions from Tunisia. J Med Virol. 2005;76(2):185-93. [PubMed ID: 15834884]. https://doi.org/10.1002/jmv.20342.
-
112.
Kchouk FH, Gorgi Y, Bouslama L, Sfar I, Ayari R, Khiri H, et al. Phylogenetic analysis of isolated HCV strains from tunisian hemodialysis patients. Viral Immunol. 2013;26(1):40-8. [PubMed ID: 23374151]. https://doi.org/10.1089/vim.2012.0043.
-
113.
Brahim I, Akil A, Mtairag el M, Pouillot R, Malki AE, Nadir S, et al. Morocco underwent a drift of circulating hepatitis C virus subtypes in recent decades. Arch Virol. 2012;157(3):515-20. [PubMed ID: 22160625]. https://doi.org/10.1007/s00705-011-1193-7.
-
114.
Rebbani K, Ouladlahsen A, Bensghir A, Akil A, Lamdini H, Issouf H, et al. Co-infections with hepatitis B and C viruses in human immunodeficiency virus-infected patients in Morocco. Clin Microbiol Infect. 2013;19(10):454-7. [PubMed ID: 23731409]. https://doi.org/10.1111/1469-0691.12252.
-
115.
Trimbitas RD, Serghini FZ, Lazaar F, Baha W, Foullous A, Essalhi M, et al. The "hidden" epidemic: a snapshot of Moroccan intravenous drug users. Virol J. 2014;11:43. [PubMed ID: 24602336]. https://doi.org/10.1186/1743-422X-11-43.
-
116.
Foullous A. Epidemiological and virological study of hepatitis c virus infection in hemodialysis (case of six centers) in morocco. J Biol Agric health. 2015;5(6):99-105.
-
117.
Alashek W, Altagdi M. Risk factors and genotypes of hepatitis C virus infection in libyan patients. Libyan J Med. 2008;3(4):162-5. [PubMed ID: 21499468]. https://doi.org/10.4176/080425.
-
118.
Elasifer HA, Agnnyia YM, Al-Alagi BA, Daw MA. Epidemiological manifestations of hepatitis C virus genotypes and its association with potential risk factors among Libyan patients. Virol J. 2010;7:317. [PubMed ID: 21073743]. https://doi.org/10.1186/1743-422X-7-317.
-
119.
Mudawi HM, Smith HM, Fletcher IA, Fedail SS. Prevalence and common genotypes of HCV infection in Sudanese patients with hepatosplenic schistosomiasis. J Med Virol. 2007;79(9):1322-4. [PubMed ID: 17607776]. https://doi.org/10.1002/jmv.20865.
-
120.
Shobokshi OA, Serebour FE, Skakni LI. Hepatitis C genotypes/subtypes among chronic hepatitis patients in Saudi Arabia. Saudi Med J. 2003;24 Suppl 2:87-91. [PubMed ID: 12897908].
-
121.
Al-Traif I, Handoo FA, Al-Jumah A, Al-Nasser M. Chronic hepatitis C. Genotypes and response to anti-viral therapy among Saudi patients. Saudi Med J. 2004;25(12):1935-8. [PubMed ID: 15711670].
-
122.
Marie MAM. Genotyping of Hepatitis C virus (HCV) in infected patients from Saudi Arabia. Afr J Microbiol Res. 2011;5(16):2388-90.
-
123.
Akbar HO, Al Ghamdi A, Qattan F, Fallatah HI, Al Rumani M. Chronic hepatitis C in saudi arabia: three years local experience in a university hospital. Hepat Mon. 2012;12(9). ee6178. [PubMed ID: 23087760]. https://doi.org/10.5812/hepatmon.6178.
-
124.
Al Traif I, Al Balwi MA, Abdulkarim I, Handoo FA, Alqhamdi HS, Alotaibi M, et al. HCV genotypes among 1013 Saudi nationals: a multicenter study. Ann Saudi Med. 2013;33(1):10-2. [PubMed ID: 23458933]. https://doi.org/10.5144/0256-4947.2013.10.
-
125.
Shier MK, El-Wetidy MS, Ali HH, Al-Qattan MM. Characterization of hepatitis C virus genotypes by direct sequencing of HCV 5'UTR region of isolates from Saudi Arabia. PLoS One. 2014;9(8). ee103160. [PubMed ID: 25099694]. https://doi.org/10.1371/journal.pone.0103160.
-
126.
Al-Kubaisy WA, Niazi AD, Kubba K. History of miscarriage as a risk factor for hepatitis C virus infection in pregnant Iraqi women. East Mediterr Health J. 2002;8(2-3):239-44. [PubMed ID: 15339110].
-
127.
Al-Kubaisy WA, Al-Naib KT, Habib MA. Prevalence of HCV/HIV co-infection among haemophilia patients in Baghdad. East Mediterr Health J. 2006;12(3-4):264-9. [PubMed ID: 17037693].
-
128.
Abdullah AM, Hardan A, Latif II. Genotyping of hepatitis C virus isolates from Iraqi hemodialysis patients by reverse transcription-PCR and one step nested RT-PCR. Diyala. 2012;3(1):9-18.
-
129.
Al-Kubaisy WA. Specific HCV antibodies, RNA, and genotypes detection correlated to the age of pregnant women in Iraq. Int J Inf Dis. 2012;16. ee67.
-
130.
Othman AA, Eissa AA, Markous RD, Ahmed BD, Al-Allawi NA. Hepatitis C virus genotypes among multiply transfused hemoglobinopathy patients from Northern Iraq. Asian J Transfus Sci. 2014;8(1):32-4. [PubMed ID: 24678171]. https://doi.org/10.4103/0973-6247.126687.
-
131.
Sharara A, Ramia S, Ramlawi F, Fares JE, Klayme S, Naman R. Genotypes of hepatitis C virus (HCV) among positive Lebanese patients: comparison of data with that from other Middle Eastern countries. Epidemiol Infect. 2007;135(3):427-32.
-
132.
Makhoul NJ, Choueiri MB, Kattar MM, Soweid AM, Shamseddeen W, Estephan HC, et al. Distribution of hepatitis C virus (HCV) genotypes among HCV infection risk groups in Lebanon. J Clin Virol. 2008;41(2):166-7. [PubMed ID: 18054277]. https://doi.org/10.1016/j.jcv.2007.10.012.
-
133.
Mahfoud Z, Kassak K, Kreidieh K, Shamra S, Ramia S. Distribution of hepatitis C virus genotypes among injecting drug users in Lebanon. Virol J. 2010;7:96. [PubMed ID: 20465784]. https://doi.org/10.1186/1743-422X-7-96.
-
134.
Bdour S. Hepatitis C virus infection in Jordanian haemodialysis units: serological diagnosis and genotyping. J Med Microbiol. 2002;51(8):700-4. [PubMed ID: 12171303]. https://doi.org/10.1099/0022-1317-51-8-700.
-
135.
Rashdan A, Hijjawi S, Jadallah K, Matalka I. Prevalence of Hepatitis C Virus Antibodies Among Blood Donors in Northern Jordan (Brief Communications). Jordan Med J. 2008;42(3).
-
136.
Al-Sweedan S, Jaradat S, Amer K, Hayajneh W, Haddad H. Seroprevalence and genotyping of hepatitis C virus in multiple transfused Jordanian patients with β-thalassemia major. Turk J Hematol. 2011;28(1):47-51. https://doi.org/10.5152/tjh.2011.05.
-
137.
Qadi AA, Tamim H, Ameen G, Bu-Ali A, Al-Arrayed S, Fawaz NA, et al. Hepatitis B and hepatitis C virus prevalence among dialysis patients in Bahrain and Saudi Arabia: a survey by serologic and molecular methods. Am J Infect Control. 2004;32(8):493-5. [PubMed ID: 15573057]. https://doi.org/10.1016/S0196655304003669.
-
138.
Abdulla MAM, Al Qamish JRA. Hepatitis C virus infection: A single center experience. Bahrain Medical Bulletin. 2008;30(1):3-8.
-
139.
Pacsa AS, Al-Mufti S, Chugh TD, Said-Adi G. Genotypes of hepatitis C virus in Kuwait. Medical Principles and Practice. 2001;10(1):55-7.
-
140.
Al-Enzi SA, Ismail WA, Alsurayei SA, Ismail E. Peginterferon alfa-2b and ribavirin therapy in Kuwaiti patients with chronic hepatitis C virus infection. East Mediterr Health J. 2011;17(8):669-78. [PubMed ID: 21977570].
-
141.
Abro AH, Al-Dabal L, Younis NJ. Distribution of hepatitis C virus genotypes in Dubai, United Arab Emirates. J Pak Med Assoc. 2010;60(12):987-90. [PubMed ID: 21381547].
-
142.
Alfaresi MS. Prevalence of hepatitis C virus (HCV) genotypes among positive UAE patients. Mol Biol Rep. 2011;38(4):2719-22. [PubMed ID: 21104026]. https://doi.org/10.1007/s11033-010-0415-5.
-
143.
Ayesh BM, Zourob SS, Abu-Jadallah SY, Shemer-Avni Y. Most common genotypes and risk factors for HCV in Gaza strip: a cross sectional study. Virol J. 2009;6:105. [PubMed ID: 19607718]. https://doi.org/10.1186/1743-422X-6-105.
-
144.
John AK, Al KS, John A, Singh R, Derbala M. Audit of state-funded antiviral treatment for chronic hepatitis C in Qatar. East Mediterr Health J. 2010;16(11):1121-7. [PubMed ID: 21218734].
-
145.
Antaki N, Haddad M, Kebbewar K, Abdelwahab J, Hamed O, Aaraj R, et al. The unexpected discovery of a focus of hepatitis C virus genotype 5 in a Syrian province. Epidemiol Infect. 2009;137(1):79-84. [PubMed ID: 18346288]. https://doi.org/10.1017/S095026880800054X.
