Abstract
Background:
Uropathogenic Escherichia coli (UPEC) with its virulence factors is the most prevalent cause of urinary tract infection (UTI).Objectives;:
This study aimed to determine the occurrence of fim, pap, sfa, and afa genes among 100 UPEC isolates collected from patients diagnosed with UTI.Materials and Methods:
A total of 100 UPEC isolates were obtained from urine samples of patients with UTI. The prevalence of 5 virulence genes encoding type 1 fimbriae (fimH), pili associated with pyelonephritis (pap), S and F1C fimbriae (sfa and foc) and afimbrial adhesins (afa) were determined through PCR method. We also investigated the phylogenetic background of all isolates. In addition, the distribution of adhesin-encoding operons between the phylogroups was assessed.Results:
The prevalence of genes encoding for fimbrial adhesive systems was 95% for fim, 57% for pap, 16% for foc, and 81% for sfa. The operons encoding for afa afimbrial adhesins were identified in 12% of isolates. The various combinations of detected genes were designated as virulence patterns. The fim gene, which occurred in strains from all phylogenetic groups (A, B1, B2, and D) was evaluated and no significant differences were found among these groups. Conversely, significant differences were observed in relation to pap, afa, foc, and sfa operons.Conclusions:
These results indicate that the PCR method is a powerful genotypic assay for the detection of adhesin-encoding operons. Thus, this assay can be recommended for clinical use to detect virulent urinary E. coli strains, as well as epidemiological studies.Keywords
Virulence Genes Adhesion Proteins-Encoding Operons Urinary Tract Infections Uropathogenic Escherichia coli
1. Background
Urinary tract infections (UTIs) are common bacterial infections associated with considerable morbidity and health care cost (1). Escherichia coli strains capable of causing disease outside the gastrointestinal tract belong to a diverse group of isolates referred to as uropathogenic E. coli (UPEC) (2). UPEC expresses a multitude of virulence factors to break the inertia of the mucosal barrier (3). The ability of these bacteria to adhere to host epithelial cells is considered a prerequisite for the establishment of infectious diseases, mainly through expression of fimbriae (4, 5). UPEC generally possesses type 1 and P fimbriae (6). Type 1 fimbriae are characterized as having the ability to agglutinate chicken and guinea pig erythrocytes in the absence of D mannose (7). They consist of a major protein, FimA, associated with ancillary proteins FimF, FimG, and the adhesin protein FimH, encoded by the fim gene cluster (8-10). This type of fimbria is common among Enterobacteriaceae, also several variants have been strongly associated with UPEC (11). Their role in infection is unclear, although it has been suggested that they may be involved in the initial stages of colonizing in the upper respiratory tract (11, 12).
Of the adhesion-encoding genes studied, pap, sfa, and afa are prevalent in E. coli strains associated with urinary tract infections (pyelonephritis) in humans (13, 14). The pap gene cluster consists of 11 genes encoding the main component of the pilus rod (PapA), which determines 11 different serogroups, and a terminally located adhesion, PapG (15, 16). The afimbrial adhesion from a pyelonephritic E. coli isolate is P-independent, X-binding adhesion, expressed by the afa-1 operon (17) mediating the specific binding to uroepithelial cell and human erythrocyte-receptors (18). The nature of the receptor on the eukaryotic cell surface is still unknown. The S fimbriae are mannose-resistant adhesions, encoded by the sfa operon of uropathogenic E. coli (9). The presence of S fimbriae is also correlated with pathogenicity of E. coli in human meningitis and septicemia (19). The distribution pattern of afimbrial adhesin, S fimbriae and group II capsule synthesis such as foc and afa in the UPEC strains is still unclear in different parts of Iran. The purpose of this study was to compare the occurrence of fim, pap, sfa, and afa genes in E. coli strains isolated from patients with urinary tract infection residing in the southeast of Iran.
2. Objectives
This study aimed to evaluate the prevalence of different operons encoding for virulence factors among E. coli strains isolated from the urine of the patients with UTI.
3. Materials and Methods
3.1. Bacterial Isolates
One hundred non-duplicate E. coli isolates were recovered by urine culture from 250 consecutive adult patients with community-acquired pyelonephritis or cystitis over a period ranging from April to September 2013 in Zabol, Iran. The isolation and identification of E. coli isolates were performed by standard bacteriological and biochemical tests using Gram staining, catalase test, indole, methyl red, Voges-Proskauer test, nitrate reduction, urease production, Simmons citrate agar, and various sugar fermentation tests (20-22). The bacteria were maintained in Tryptic Soy Broth (TSB) (Sigma; The USA) with Glycerol at -70°C.
3.2. Clinical Data
The diagnosis of acute pyelonephritis and cystitis was based on the cytobacteriologic examination of urine and clinical investigation. UTI was defined as the presence of a positive urine culture (≥ 105 colony-forming units (cfu)/mL) and pyuria (≥ 104 leukocytes/mL of clean voided urine) (13). Diagnostic criteria for acute pyelonephritis were dysuria, temperature of ≥ 38.5°C, leukocyturia of > 105 mL, and no other identifiable source of infection (23).
3.3. Bacterial Culture and DNA Extraction
DNA extraction was performed using an optimized boiling method. E. coli strains were grown in Luria-Bertani (LB) Broth (Lonza; The USA) at 37°C overnight. Bacteria were pelleted from 1.5 mL LB broth and suspended in 200 ul of sterile distilled water, then incubated at 100°C for 10 minutes and centrifuged. One hundred microliters of the supernatant was stored at -20°C as a template DNA stock (13).
3.4. Polymerase Chain Reaction Amplification
Specific primers were used to amplify sequences of the fim, pap, sfa/foc, and afa operons. Details of primer sequences, predicted sizes of the amplified products, and specific annealing temperatures are shown in Table 1. Detection of adhesin-encoding operons (pap, sfa, and afa) and fim sequences was done by multiplex PCR.
Primers and Cycling Conditions Used for Amplification of Fimbriae Genes
| Gene | Sequence (5’ to 3’) | Size, bp | Cycling Conditions | Reference |
|---|---|---|---|---|
| fim | GTTGTTCTGTCGGCTCTGTC TAAATGTCGCACCATCCAG | 400 | 95°C for 4 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 40 s; 72°C for 3 min | (24) |
| pap | GACGGCTGTACTGCAGGGTGTGGCG ATATCCTTTCTGCAGGGATGCAATA | 328 | 95°C for 4 min; 35 cycles of 95°C for 60 s, 65°C for 60 s, 72°C for 60 s; 72°C for 2 min | (25) |
| sfa | CCGTAAAGATGTCTGCGAG AGCAAGTCTGGCAACGAG | 100 | 95°C for 4 min; 35 cycles of 95°C for 30 s, 53°C for 30 s, 72°C for 40 s; 72°C for 3 min | (24) |
| foc | GGTGGAACCGCAGAAAATAC GAACTGTTGGGGAAAGAGTG | 388 | 95°C for 4 min; 35 cycles of 95°C for 60 s, 58°C for 60 s, 72°C for 120 s; 72°C for 10 min | (26) |
| afa | GCTGGGCAGCAAACTGATAACTCTC CATCAAGCTGTTTGTTCGTCCGCCG | 750 | 95°C for 5 min; 35 cycles of 95°C for 60 s, 60°C for 30 s, 72°C for 180 s; 72°C for 7 min | (25) |
The reactions (25 μL) consisted of 10-pmol/L of each primer, 2 μL templates DNA, and 12.5 μL of a ready-to-use 2X PCR Master Mix Red (Ampliqon; Denmark) (27), with the following amplification conditions: an initial denaturation at 94°C for 10 minutes, followed by 35 DNA cycles of denaturation at 94°C for 2 minutes, annealing at a specific temperature for 30 seconds (Table 1), and extension at 72°C for 1 minute. A 5 µL aliquot of the PCR product underwent gel electrophoresis on agarose 2%, followed by staining with ethidium bromide solution. Amplified DNA elements of specific sizes were detected by UV-induced fluorescence and the size of the amplicons was estimated by comparing them with the 1 kb DNA ladder (Promega; Madison, WI, the United States of America) included on the same gel (Figure 1). The phylogenetic group to which the E. coli strains belonged was determined by a PCR-based method as described previously (28). The data of the 3 amplifications resulted in the assignment of the strains to phylogenetic groups as follows: chuA+, yjaA+, group B2; chuA+, yjaA-, group D; chuA-, TspE4.C2+, groupB1; chuA-, TspE4.C2-, group A (28).
Multiplex- PCR profiles specific for E. coli phylogenetic groups and detection of virulence genes.
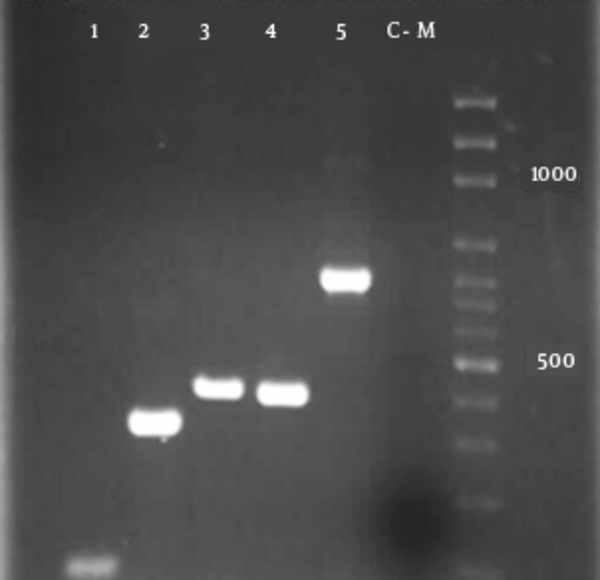
4. Results
The frequencies of the studied virulence genes are reported in Tables 2 and 3. With regard to adhesin virulence determinants, fim gene was the most common virulence gene and detected in 95% (95 out of 100) of the UTI isolates. Next, pap gene was present in 57% (57 out of 100) of isolates and sfa gene in 81% of isolates. Finally, afa and foc genes were found in 12% and 16% of isolates, respectively. One isolate was negative for all virulence genes. Based on the distribution of the various targeted sequences, all studied strains exhibited 16 virulence gene patterns, referred to as Ec (Table 2). Ec5 was characterized by the presence of fim operon only, and was the most noted pattern, found in 8 isolates. However, among 95% of isolates that were fim-positive, 2% harbored the foc gene and the other 85% exhibited distinct diversity of gene patterns (Ec1 - 4, Ec6, Ec8 - 9, and Ec12 - 15). Out of 100 UPEC isolates tested by PCR, 36% carried sequences related to the 3 adhesion-encoding (fim, sfa, and pap) operon families.
When the strains recovered from all the populations were considered, B2 and D group strains were the most common (55 and 22%, respectively), followed by A group strains (17%). B1 group strains were rare (6%) (Table 3). Most of the known extraintestinal VFs (pap, sfa, and fim genes) were concentrated within groups B2 or D, whereas foc gene was more broadly distributed between phylogenetic groups B2 and A (Table 3). Of the isolates tested, a few isolates belonged to either phylogenetic group (A, B1, and D) were positive for pap, afa, foc and sfa operons, compared to phylogenetic group B2. Evaluation of the foc operon indicated its presence in 1% and 16% of isolates belonged to phylogenetic groups A and B2, respectively, but it was not observed in phylogenetic groups B1 and D isolates (Table 3).
Virulence Patterns Identified Among the Studied Strains
| Pattern | Virulence Gene | ||||||
|---|---|---|---|---|---|---|---|
| fim | sfa | pap | foc | afa | No. of Strains | Phylogenetic Group | |
| Ec1 | + | + | + | + | + | 1 | B2 |
| Ec2 | + | + | + | + | - | 9 | B2 |
| Ec3 | + | + | + | - | - | 36 | A, B1, B2, and D |
| Ec4 | + | + | - | - | - | 21 | A, B1, B2, and D |
| Ec5 | + | - | - | - | - | 8 | A, B1, B2, and D |
| Ec6 | + | - | + | + | - | 1 | B2 |
| Ec7 | - | + | + | + | + | 2 | B2 and A |
| Ec8 | + | + | + | - | + | 2 | B2 and D |
| Ec9 | + | + | - | + | - | 3 | B2 |
| Ec10 | + | - | + | - | - | 5 | B2 and D |
| Ec11 | - | + | - | - | + | 2 | A and D |
| Ec12 | + | + | - | - | + | 5 | B2, D, and A |
| Ec13 | + | - | - | + | - | 2 | B2 and A |
| Ec14 | + | - | - | - | + | 1 | B2 |
| Ec15 | + | - | + | - | + | 1 | B2 |
| Ec16 | - | - | - | - | - | 1 | A |
| Total | 95 | 81 | 57 | 16 | 12 | 100 | |
Phylogenetic Distribution of Virulence Genes
| Ph. Group (N) | Virulence Gene | ||||
|---|---|---|---|---|---|
| Fim | sfa | pap | foc | afa | |
| A (17) | 14 | 12 | 2 | 1 | 2 |
| B1 (6) | 5 | 5 | 3 | 0 | 0 |
| B2 (55) | 55 | 46 | 46 | 16 | 5 |
| D (22) | 21 | 18 | 6 | 0 | 5 |
| TOTAL (100) | 95 | 81 | 57 | 17 | 12 |
5. Discussion
Escherichia coli causes the vast majority of UTIs in both ambulatory and hospitalized patients (29). The degree of severity depends on the virulence of the responsible strains and susceptibility of the host, particularly if there is a concomitant urological illness. A better knowledge of the virulence characteristics of the microorganism causing the infection will allow the clinician to anticipate, up to a point, the evolution of infection in the patient. To the best of our knowledge, our study was the first to demonstrate associations between E. coli adhesin-encoding operons and UTI in Iran. Several virulence determinants contribute to the pathogenicity of E. coli in UTI (17, 19, 27). They are the product of different genes, which can be detected by PCR method (9, 22). However, there is always the possibility of mutation at the level of the corresponding gene, leading to the absence of its detection. Therefore, a positive PCR shows the presence of the virulence gene, but a negative PCR does not rule out the presence of the corresponding operon. However, this phenomenon remains scarce.
Genes coding for adhesive systems represent the most common factors for the virulence of E. coli in UTI. The distribution of adhesin-encoding operons in our strains was in agreement with other published data (30-32). The present data indicate the crucial role of these adhesin-encoding operons in E. coli-associated UTI (33, 34). Moreover, an important role of pap adhesion genes in the pathophysiology of UTIs caused by E. coli has been reported in several studies (32, 35). In addition, the presence of foc and afa virulence genes was 16% and 12%, respectively. The presence of combined pap and afa virulence genes was observed in 50% of isolates, indicating high presence of virulence genes in isolates collected from patients with UTIs in Iran as described (32, 36). The presence of higher number of observed combined pap and sfa genes was in accordance with other reports (30-32). The higher number of isolates having pap and sfa genes together as compared to other combinations could be due to the localization of these genes on the same pathogenicity island of UPEC strains (37). Our results show a higher frequency of fim operon compared to the rest of the genes, which may indicate a crucial role of the virulence genes in E. coli causing UTI.
Regarding P fimbriae, our results are inconsistent with those of many studies, indicating that among patients with UTIs, 57% possess P fimbriae (13, 32). The difference of occurrence in the percentage of the structural adhesins (P fimbriae) may be due to the different environmental niches prevailing at the two different host sites, as described previously (38). In contrast to other studies, the sfa operon was observed in 80% in UTIs isolates (13, 35, 39). This may indicate that sfa operon plays an important role in causing UTI in Zabol, southeast of Iran. In addition, we propose a more significant role for fim, pap, and sfa operons in the generation of UTI. Based on the distribution of the various target sequences, the strains studied exhibited 16 most common virulence patterns, referred to as Ec followed by an Arabic numeral (Table 2). The UTIs isolates exhibited a great diversity of gene patterns, showing Ec1, Ec2, Ec3, and Ec16 patterns that were in agreement with other report (13, 40). The presence of the foc operon together with the pap operon was detected in the same strains as the Ec1, Ec2, Ec6, and Ec9 pattern (Table 2); this association had been previously reported (13). A codependence of these virulence factors in a particular pathogenic pathway has been discussed (40) but needs to be confirmed.
When phylogenetic analysis of these virulence factors is done, a striking difference was observed between the foc operon on one hand, and the other pathogenic determinants, including the fim, sfa, and pap operon, on the other hand. The foc operon is strictly restricted to strains of the phylogenetic B2 group (16 out of 17) and phylogenetic A group (1 out of 17) (Table 3). Similarly, Maslow et al. (41) reported that within adult bloodstream isolates, the presence of this operon was restricted to one cluster. In contrast, the other pathogenic determinants, although being predominant in the B2 group, are also distributed among the other phylogenetic groups. It can be proposed that most of the genes needed for causing neonatal meningitis belonged to the E. coli B2 phylogenetic group initially and horizontal transfer of these genes has occurred toward the more genetically distant groups.
In conclusion, our study showed that: 1) a high prevalence of fin, pap, and sfa operon may be responsible for UTIs, 2) the characterization of E. coli strains isolated from UTI is of great interest to improve our knowledge regarding their virulence genetic determinants, (3) further studies are needed to identify E. coli virulence factors responsible for UTI and to determine the physiopathology of these infections to consider possible preventive measures.
Acknowledgements
References
-
1.
Agarwal J, Srivastava S, Singh M. Pathogenomics of uropathogenic Escherichia coli. Indian J Med Microbiol. 2012;30(2):141-9. [PubMed ID: 22664427]. https://doi.org/10.4103/0255-0857.96657.
-
2.
Wang X, Yan Q, Xia X, Zhang Y, Li D, Wang C, et al. Serotypes, virulence factors, and antimicrobial susceptibilities of vaginal and fecal isolates of Escherichia coli from giant pandas. Appl Environ Microbiol. 2013;79(17):5146-50. [PubMed ID: 23793635]. https://doi.org/10.1128/AEM.01367-13.
-
3.
Bien J, Sokolova O, Bozko P. Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. Int J Nephrol. 2012;2012:681473. [PubMed ID: 22506110]. https://doi.org/10.1155/2012/681473.
-
4.
Beachey EH. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J Infect Dis. 1981;143(3):325-45. [PubMed ID: 7014727].
-
5.
Ike K, Kume K, Kawahara K, Danbara H. Serotyping of O and pilus antigens of Escherichia coli strains isolated from chickens with coli-septicemia. Nihon Juigaku Zasshi. 1990;52(5):1023-7. [PubMed ID: 1704075].
-
6.
Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect Immun. 2006;74(2):1387-93. [PubMed ID: 16428790]. https://doi.org/10.1128/IAI.74.2.1387-1393.2006.
-
7.
Kariyawasam S, Nolan LK. Pap mutant of avian pathogenic Escherichia coli O1, an O1:K1:H7 strain, is attenuated in vivo. Avian Dis. 2009;53(2):255-60. [PubMed ID: 19630233].
-
8.
Orndorff PE, Falkow S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J Bacteriol. 1984;159(2):736-44. [PubMed ID: 6146599].
-
9.
Stordeur P, Bree A, Mainil J, Moulin-Schouleur M. Pathogenicity of pap-negative avian Escherichia coli isolated from septicaemic lesions. Microbes Infect. 2004;6(7):637-45. [PubMed ID: 15158770]. https://doi.org/10.1016/j.micinf.2004.03.006.
-
10.
Feria C, Machado J, Correia JD, Goncalves J, Gaastra W. Virulence genes and P fimbriae PapA subunit diversity in canine and feline uropathogenic Escherichia coli. Vet Microbiol. 2001;82(1):81-9. [PubMed ID: 11423198].
-
11.
Simms AN, Mobley HL. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J Bacteriol. 2008;190(10):3747-56. [PubMed ID: 18359812]. https://doi.org/10.1128/JB.01870-07.
-
12.
Katouli M. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J Microbiol. 2010;2(2):59-72. [PubMed ID: 22347551].
-
13.
Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17(6):e450-3. [PubMed ID: 23510539]. https://doi.org/10.1016/j.ijid.2013.01.025.
-
14.
Girardeau JP, Lalioui L, Said AM, De Champs C, Le Bouguenec C. Extended virulence genotype of pathogenic Escherichia coli isolates carrying the afa-8 operon: evidence of similarities between isolates from humans and animals with extraintestinal infections. J Clin Microbiol. 2003;41(1):218-26. [PubMed ID: 12517852].
-
15.
Brunder W, Khan AS, Hacker J, Karch H. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(-). Infect Immun. 2001;69(7):4447-57. [PubMed ID: 11401985]. https://doi.org/10.1128/IAI.69.7.4447-4457.2001.
-
16.
Srimanote P, Paton AW, Paton JC. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2002;70(6):3094-100. [PubMed ID: 12011003].
-
17.
Labigne-Roussel A, Falkow S. Distribution and degree of heterogeneity of the afimbrial-adhesin-encoding operon (afa) among uropathogenic Escherichia coli isolates. Infect Immun. 1988;56(3):640-8. [PubMed ID: 2893773].
-
18.
Shetty AV, Kumar SH, Shekar M, Shetty AK, Karunasagar I, Karunasagar I. Prevalence of adhesive genes among uropathogenic Escherichia coli strains isolated from patients with urinary tract infection in Mangalore. Indian J Med Microbiol. 2014;32(2):175-8. [PubMed ID: 24713907]. https://doi.org/10.4103/0255-0857.129812.
-
19.
Antao EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 2009;1(1):22. [PubMed ID: 20003270]. https://doi.org/10.1186/1757-4749-1-22.
-
20.
Perfeito R, Cunha-Oliveira T, Rego AC. Reprint of: revisiting oxidative stress and mitochondrial dysfunction in the pathogenesis of Parkinson disease-resemblance to the effect of amphetamine drugs of abuse. Free Radic Biol Med. 2013;62:186-201. [PubMed ID: 23743292]. https://doi.org/10.1016/j.freeradbiomed.2013.05.042.
-
21.
Falakian Z, Nikookar, Nafisi M, Karimi A, Validi M. Frequency of Class 1 Integrons among Escherichia coli Isolates of Patients with Urinary Tract Infection. Arch Clin Infect Dis. 2012;6(4):157-60.
-
22.
Goudarzi H, Aghamohammad S, Hashemi A, Nikmanesh B, Noori M. Distribution of blaTEM, blaSHV and blaCTX-M Genes Among Escherichia coli Isolates Causing Urinary Tract Infection in Children. Arch Clin Infect Dis. 2013;8(3). eee16207. https://doi.org/10.5812/archcid.16207.
-
23.
Sakran W, Smolkin V, Odetalla A, Halevy R, Koren A. Community-acquired urinary tract infection in hospitalized children: etiology and antimicrobial resistance. A comparison between first episode and recurrent infection. Clin Pediatr (Phila). 2015;54(5):479-83. [PubMed ID: 25385933]. https://doi.org/10.1177/0009922814555974.
-
24.
Nazemi A, Naderi M, Jafarpour M, Mirinargesi M, Sharifi SH. The Detection of Fimbrial Pathogenic Genes in E. coli Strains Isolated from Patients with Urinary Tract Infection. Med Lab J. 2010;4(2):31-7.
-
25.
Birosova E, Siegfried L, Kmet'ova M, Makara A, Ostro A, Gresova A, et al. Detection of virulence factors in alpha-haemolytic Escherichia coli strains isolated from various clinical materials. Clin Microbiol Infect. 2004;10(6):569-73. [PubMed ID: 15191387]. https://doi.org/10.1111/j.1469-0691.2004.00922.x.
-
26.
Mitsumori K, Terai A, Yamamoto S, Yoshida O. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol Med Microbiol. 1998;21(4):261-8. [PubMed ID: 9752998].
-
27.
Rashki A. Cervico-vaginopathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Microb Pathog. 2014;75:29-34. [PubMed ID: 25193497]. https://doi.org/10.1016/j.micpath.2014.08.004.
-
28.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555-8. [PubMed ID: 11010916].
-
29.
Srinivasan U, Foxman B, Marrs CF. Identification of a gene encoding heat-resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J Clin Microbiol. 2003;41(1):285-9. [PubMed ID: 12517862].
-
30.
Bogyiova E, Kmetova M, Biros E, Siegfried L. Detection of pap-, sfa- and afa-specific DNA sequences in Escherichia coli strains isolated from extraintestinal material. Folia Microbiol (Praha). 2002;47(6):723-6. [PubMed ID: 12630326].
-
31.
Becker Saidenberg A, Robaldo Guedes NM, Fernandes Seixas GH, da Costa Allgayer M, Pacifico de Assis E, Fabio Silveira L, et al. A Survey for Escherichia coli Virulence Factors in Asymptomatic Free-Ranging Parrots. ISRN Vet Sci. 2012;2012:984813. [PubMed ID: 23738135]. https://doi.org/10.5402/2012/984813.
-
32.
Mohajeri P, Khademi H, Ebrahimi R, Farahani A, Rezaei M. Frequency distribution of virulence factors in uropathogenic Escherichia coli isolated from Kermanshah in 2011-2012. Int J Appl Basic Med Res. 2014;4(2):111-6. [PubMed ID: 25143887]. https://doi.org/10.4103/2229-516X.136794.
-
33.
Garcia MI, Gounon P, Courcoux P, Labigne A, Le Bouguenec C. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol Microbiol. 1996;19(4):683-93. [PubMed ID: 8820639].
-
34.
Usein CR, Damian M, Tatu-Chitoiu D, Capusa C, Fagaras R, Tudorache D, et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J Cell Mol Med. 2001;5(3):303-10. [PubMed ID: 12067489].
-
35.
Farshad S, Emamghorashi F. The prevalence of virulence genes of E. coli strains isolated from children with urinary tract infection. Saudi J Kidney Dis Transpl. 2009;20(4):613-7. [PubMed ID: 19587502].
-
36.
Karimian A, Momtaz H, Madani M. Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr J Microbiol Res. 2012;6(39):6811-6.
-
37.
Dobrindt U, Blum-Oehler G, Hartsch T, Gottschalk G, Ron EZ, Funfstuck R, et al. S-Fimbria-encoding determinant sfa(I) is located on pathogenicity island III(536) of uropathogenic Escherichia coli strain 536. Infect Immun. 2001;69(7):4248-56. [PubMed ID: 11401961]. https://doi.org/10.1128/IAI.69.7.4248-4256.2001.
-
38.
Moulin-Schouleur M, Reperant M, Laurent S, Bree A, Mignon-Grasteau S, Germon P, et al. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol. 2007;45(10):3366-76. [PubMed ID: 17652485]. https://doi.org/10.1128/JCM.00037-07.
-
39.
Mercon M, Regua-Mangia AH, Teixeira LM, Irino K, Tuboi SH, Goncalves RT, et al. Urinary tract infections in renal transplant recipients: virulence traits of uropathogenic Escherichia coli. Transplant Proc. 2010;42(2):483-5. [PubMed ID: 20304171]. https://doi.org/10.1016/j.transproceed.2010.01.049.
-
40.
Graciano MF, Santos LR, Curi R, Carpinelli AR. NAD(P)H oxidase participates in the palmitate-induced superoxide production and insulin secretion by rat pancreatic islets. J Cell Physiol. 2011;226(4):1110-7. [PubMed ID: 20857410]. https://doi.org/10.1002/jcp.22432.
-
41.
Maslow JN, Whittam TS, Gilks CF, Wilson RA, Mulligan ME, Adams KS, et al. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63(7):2409-17. [PubMed ID: 7790051].