Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria
- Published
- Accepted
- Received
- Academic Editor
- Pedro Silva
- Subject Areas
- Genetics, Microbiology, Infectious Diseases
- Keywords
- Resistance genes, Chicken, Susceptibility, Antibiotics, Pathogens
- Copyright
- © 2018 Akinbami et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria. PeerJ 6:e5053 https://doi.org/10.7717/peerj.5053
Abstract
A serious concern is arising on the coexistence of extended-spectrum beta-lactamase (ESBL) and plasmid mediated quinolone resistance (PMQR) producing bacteria in animal husbandry, which could be transferred to humans, especially in strains that may not be routinely screened for resistance. This study therefore tested the prevalence of ESBL and PMQR genes in selected bacteria isolated from poultry faeces. Faecal droppings of birds were collected from 11 farms in five states in South Western Nigeria. Bacteria were isolated from the samples on cefotaxime supplemented plates and identified with MALDI-TOF. The MIC was determined using VITEK system and resistance genes were detected with PCR. A total of 350 strains were isolated from different samples and selected strains were identified as 23 Klebsiella pneumonia, 12 Morganella morganii, seven Leclercia adecarboxylata and one Citrobacter freundii. All the species were resistant to gentamycin, trimethoprim/sulphamethaxole, tobramycin, piperacillin, cefotaxime and aztreonam (except Morganella morganii strains which were mostly susceptible to aztreonam). All the tested strains were susceptible to imipenem, meropenem and amikacin. All Leclercia adecarboxylata strains were resistant to ceftazidime, cefepime and fosfomycin while all Morganella morganii strains were resistant to fosfomycin, moxifloxacin and ciprofloxacin. All tested species were generally sensitive to ciprofloxacin except Morganella morganii strains which were resistant to ciprofloxacin. The resistance to ciprofloxacin, ceftazidime, cefepime, tigercylin, colistin and fosfomycin were 65%, 40%, 23%,, 7%, 33%, 48% respectively while the prevalence of SHV, TEM and CTX genes were 42%, 63%, 35% respectively. 9.3% of the isolates had the three ESBL genes, 2.33% had qnrA gene, 4.65% had qnr B gene while none had qnrS gene. The most prevalent PMQR gene is Oqxb (25.58%) while 6.98% had the qep gene. Klebsiella pneumoniae generally had both ESBL and PMQR genes. The high prevalence of extended spectrum beta-lactamase genes in the studied strains calls for caution in the use of beta lactam antibiotics in poultry feeds. This is the first report of the occurrence of extended spectrum beta-lactamase and plasmid mediated quinolone resistance genes in Morganella morganii and Leclercia adecarboxylata strains isolated from poultry faeces.
Introduction
The use of sub-therapeutic antibiotics as growth promoters in poultry feeds in some countries has led to increasing rate of resistance to antibiotics among pathogens in poultry environment. This is enhanced by the ability of the resistant strains to transfer acquired resistance to their progeny and other unrelated bacteria through plasmids (Apata, 2009). Resistance is common to the most frequently prescribed antibiotics such as the quinolones, beta-lactam antibiotics and aminoglycosides. This is observed in different types of infections caused by Gram-negative bacteria, especially Enterobacteriaceae, which may be difficult to treat (Schultsz & Gerlings, 2012). Of more concern are quinolones and beta-lactam antibiotics, which are commonly prescribed in human to treat different infections.
Resistance to quinolones varies across microorganisms and geographical locations. There seems to be a linkage between resistance to quinolones and the β-lactam antibiotics as ESBL genes are frequently carried on plasmids which can also carry genes encoding resistance to other antibiotics e.g., aminoglycosides thereby giving ESBL producers the characteristic broad antibiotic resistance to multiple antibiotic classes (Rawat & Nair, 2010). ESBLs are most commonly found in Enterobacteriaceae and one of the commonest ESBL producers is Escherichia coli (Mansouri & Ramazanzadeh, 2009). They could also occur in uncommon species of Enterobacteriaceae e.g., Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii. Morganella morganii could cause nosocomial and opportunistic infections in intensive care unit patients and immunocompromised hosts (Singla et al., 2010). It has been reported to cause fatal infections in chicken (Zhao et al., 2012). Leclercia adecarboxylata is another rare Enterobacteriaceae isolated from water, which could act as an opportunistic pathogen in immunocompromised patients. It is usually susceptible to most commonly used antibiotics including beta-lactams. However, a few cases of antibiotic-resistant L. adecarboxylata have been reported (Shin et al., 2012). Citrobacter freundii is resistant to β-lactam antibiotics due to the production of ESBL in some strains (Kanamori et al., 2011).
Escherichia coli and Salmonella spp. are often isolated in poultry and its environment as ESBL producers (Saliu, Vahjen & Zentek, 2017) while E. coli and Enterobacter cloacae were the most frequently identified organisms from hatching eggs (Mezhoud et al., 2016). There are information on ESBL-producing E. coli strains from poultry, but little information is available on detection of ESBL-producing Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii in poultry. We have previously studied susceptibility of non E. coli enterobacteriaceae isolated from poultry in Ibadan, Nigeria but the strains were not identified to species level, neither were resistant genes investigated (Ayeni, Olujobi & Alabi 2015). The aim of this study was therefore to determine the presence of ESBL and PMQR resistance genes in Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii strains isolated from faecal dropping of birds in 11 poultry farms from South Western Nigeria.
Materials and Methods
Isolation procedures
The samples were collected between September 2015 and February 2016. A total of 240 fecal droppings were collected aseptically from 11 commercial poultry farms from five states in South Western Nigeria i.e., Oyo, Ondo, Ogun, Ekiti and Osun. Immediately after collection, 1g of each fecal sample was homogenized in 9 mL of MacConkey broth (Oxoid, Cheshire, UK) supplemented with cefotaxime (2 µg/mL) and incubated at 37 °C for 24 h. The incubated broth was plated on MacConkey agar plates (Oxoid, Cheshire, UK) supplemented with cefotaxime (2 µg/mL) (Tamang et al., 2013) at appropriate dilutions and incubated at 37 °C for 24 h. All possible distinct colonies with different morphology that grew on MacConkey plates were sub cultured to get pure cultures which were stored appropriately.
Identification of isolates
Overnight cultures of bacterial isolates were identified with MALDI-TOF apparatus (VITEK® MS (Biomerieux, Nuertingen, Germany)) according to the manufacturer’s instructions. In summary, bacterial strains were grown on MacConkey agar plates for 24 h. A thin smear of the strains were made on MALDI plate and overlaid with 1 µL of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid) and air dried at room temperature. The plates were put in VITEK® MS (Biomerieux, Nuertingen, Germany) where the organisms were identified by comparing their mass spectra with reference spectra of the manufacturer database. Data were interpreted with scores of ≥2 considered as reliable species level identification. All identified Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii were selected for further studies.
Antibiotic susceptibility of isolates
Bacterial isolates were cultured on MacConkey agar plates and incubated overnight aerobically. Appropriate dilutions of the colonies were made and put into VITEK apparatus for MIC evaluation. The MIC values of different classes of antibiotics were determined by VITEK®2 compact system (AST-N248 cards; Biomerieux, Nuertingen, Germany) according to the manufacturer’s instructions.
Detection of ESBL and PMQR genes
All the isolates were subcultured on MacConkey agar and incubated for 24 hrs. Two colonies of grown bacteria were put in an eppendorf tube containing 1 mL of molecular grade water and mixed with the aid of a vortex mixer. The mixture was boiled for 10 min in a water bath at 100 °C and afterwards centrifuged for 5 min at 1,000 rpm. The supernatant (containing the DNA) was removed carefully using a micropipette without disturbing the pellet. The DNA was stored at −20 °C for PCR analysis.
The primers and PCR conditions used in this study are shown in Table 1. The primers were synthesized by Inqaba Biotechnical Industries (Pty) Ltd, Hatfield, South Africa. Positive and negative control from our laboratory were used. All PCR mixture contained the master mix (half of the total reaction volume), primers (1% of the final volume of the supermix), molecular graded water and 1 µL of the DNA template. The final reaction volume was 20 µl. The PCR product was thereafter viewed by gel electrophoresis after staining in ethidium bromide. A band corresponding to the expected size and positive control was assessed as positive.
| Gene | Primers | Primer sequence | Annealing temp. | Product size | References |
|---|---|---|---|---|---|
| qnrA | Qnr(A)-F | 5-ATTTCTCACGCCAGGATTTG-3 | 53 °C | 516 bp | Wang et al. (2009) |
| Qnr(A)-R | 5-GATCGGCAAAGGTTAGGTCA-3 | ||||
| qnrB | Qnr(B)-F | 5-GATCGTGAAAGCCAGAAAGG-3 | 53 °C | 469 bp | Wang et al. (2009) |
| Qnr(B)-R | 5-ACGATGCCTGGTAGTTGTCC-3 | ||||
| qnrS | Qnr(S)-F | 5-ACGACATTCGTCAACTGCAA-3 | 53 °C | 417 bp | Wang et al. (2009) |
| Qnr(S)-R | 5-TAAATTGGCACCCTGTAGGC-3 | ||||
| qepA | Qep(A)-F | 5-CTTCTCTGGATCCTGGACAT-3 | 53 °C | 720 bp | Şahinturk et al. (2016) |
| Qep(A)-R | 5-TGAAGATGTAGACGCCGAAC-3 | ||||
| oqxB | Oqx(B)-F | 5-ATCGGTATCTTCCAGTCACC-3 | 56 °C | 541 bp | Şahinturk et al. (2016) |
| Oqx(B)-R | 5-ACTGTTTGTAGAACTGGCCG-3 | ||||
| SHV | SHV-F | 5-TCGCCTGTGTATTATCTCCC-3 | 50 °C | 768 bp | Maynard et al. (2003) |
| SHV-R | 5-CGCAGATAAATCACCACAATG-3 | ||||
| TEM | TEM-F | 5-GAGTATTCAACATTTTCGT-3 | 50 °C | 857 bp | Maynard et al. (2003) |
| TEM-R | 5-ACCAATGCTTAATCAGTGA-3 | ||||
| CTX-M | CTX-F | 5-TTTGCGATGTGCAGTACCAGT AA-3 | 56 °C | 543 bp | Edelstein et al. (2003) |
| CTX-R | 5-CGATACGTTGGTGGTGCCATA-3 |
Results
A total of 350 strains were isolated from different samples and identified. The prevalence of the studied enterobacteriaceae were 7% Klebsiella pneumonia (23 strains isolated from four farms), 3% Morganella morganii (12 strains isolated from one farm), 2% Leclercia adecarboxylata (seven strains isolated from two farms) and 0.3% Citrobacter freundii. The remaining 308 strains were strains of E. coli and Enterobacter cloaceae.
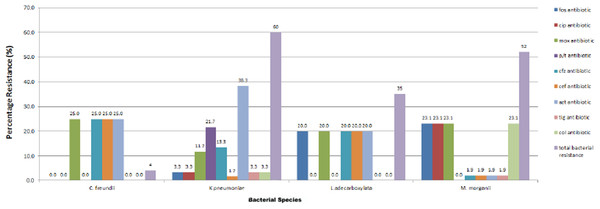
All the four species were resistant to gentamycin, trimethoprim/sulphamethaxole, tobramycin, piperacillin, cefotaxime and aztreonam (except Morganella morganii, which were mostly susceptible to aztreonam). All the strains were sensitive to imipenem, meropenem and amikacin. Most of the strains were sensitive to tigercycline. All Leclercia adecarboxylata strains were resistant to moxifloxacin, ceftazidime, cefepime and fosfomycin. All Morganella morganii strains were resistant to colistin, fosfomycin, moxifloxacin and ciprofloxacin. The only Citrobacter freundii strain had additional resistance to ceftazidime and cefepime. The tested species were mostly sensitive to ciprofloxacin but all Morganella morganii strains were resistant to ciprofloxacin. 65% of the isolates were resistant to ciprofloxacin, 40% were resistant to ceftazidime, 23% were resistant to cefepime, 7% were resistant to tigercylin, 33% were resistant to colistin while 48% were resistant to fosfomycin (Fig. 1).
Figure 1: Resistance profile of bacterial strains.
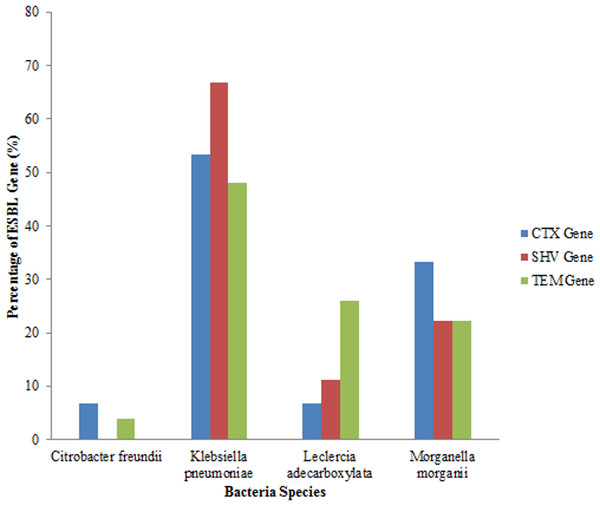
An analysis of all the isolates revealed that 18 (41.86%) had the SHV gene out of which 12 (66.67%) were Klebsiella pneumoniae, (four) 22.22% were Morganella morganii and two (11.11%) were Leclerchia adecarboxylata. Twenty seven (62.79%) of the total isolates had the TEM gene out of which 13 (48.14%) were Klebsiella pneumoniae, 7 (25.93%) were Leclerchia adecarboxylata, six (22.22%) were Morganella morganii and 1 strain of Citrobacter freundii. Also, 15 (34.88%) of the total isolates had the CTX-M gene with 8 (53.33%) being Klebsiella pneumoniae, one (6.67%) was Leclerchia adecarboxylata, 5 (33.33%) were Morganella morganii and 6.67% was one isolate of Citrobacter freundii. Fifteen (34.88%) isolates had both the SHV and TEM genes while four (9.3%) isolates had the three genes and seven (16.28%) isolates had none of the genes. All the Leclerchia adecarboxylata species had the SHV gene while Klebsiella pneumoniae had more SHV and TEM genes than CTX-M gene (Fig. 2).
Figure 2: Distribution of ESBL genes in bacterial strains.
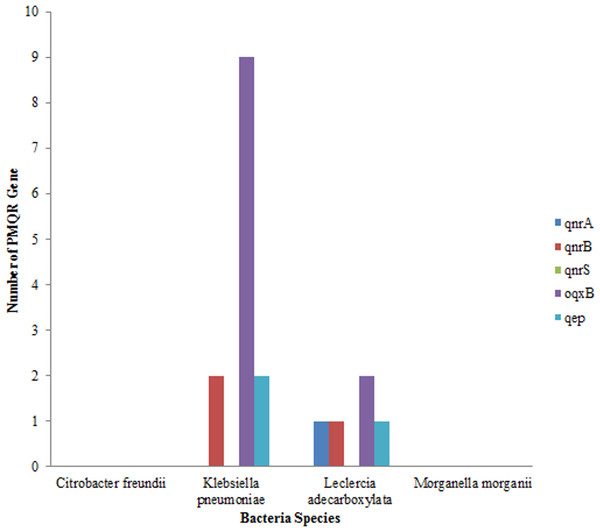
Out of the total of 43 isolates, qnr A gene was only detected in one isolate of Leclerchia adecarboxylata, only two isolates of Klebsiella pneumoniae had the qnr B, gene while no isolate had the qnr S gene. The most prevalent PMQR gene was Oqxb in 11 strains (nine isolates of Klebsiella pneumoniae and two isolates of Leclerchia adecarboxylata). In addition, two isolates of Klebsiella pneumoniae and one isolate of Leclerchia adecarboxylata had the qep gene. None of the isolates had all the PMQR genes (Fig. 3).
Figure 3: Distributions of PMQR among bacterial strains.
Discussion
Antibiotic resistance has become a menace in the environment, as it has rendered the use of several antibiotics in the treatment of certain infections ineffective. A high prevalence of resistant bacteria genes in poultry environment may increase the risk of spread to humans, as the poultry environment is constantly in contact with humans. Observed total resistance of all studied strains to cefotaxime, piperacillin, tobramycin, trimethoprim/sulfamethoxazole and gentamicin conforms to the study by Bradford (2001), in which it was reported that β-lactam antibiotics such as ceftazidime, cefotaxime and oxyimino-monobactam were ineffective against studied bacteria however, carbapenems and cephamycine were effective against ESBL producing strains (Seija et al., 2015). This was also observed in this study, with total susceptibility of all tested organisms to amikacin, imipenem and meropenem. Carbapenems are effective in the treatment of infections caused by multi-drug resistant Gram-negative bacteria including those that produce ESBLs (Walsh et al., 2005). All Morganella morganii and Leclercia adecarboxylata strains in this study were resistant to fosfomycin. It has also been previously noted that ESBL producing isolates displayed higher fosfomycin resistance than ESBL negative strains (Seija et al., 2015). The total resistance of Morganella morganii strains to fosfomycin, colistin and ciprofloxacin is alarming. Demir & Buyukguclu (2013) has also previously reported resistance of all Morganella morganii strains from urogenital infections to fosfomycin and Arca, Reguera & Hardisson (1997) has also reported incorporation of alpha-glycerolphosphate in a fosfomycin resistant Morganella morgannii strain. However, Seija et al. (2015) have successfully reported treatment of multidrug resistant Morganella morganii in sepsis with fosfomycin thereby suggesting that the resistance is not intrinsic. Resistance of Morganella morganii to colistin has been described as intrinsic (Liu et al., 2016).
TEM gene was the most prevalent ESBL gene detected followed by SHV and CTX-M gene in clinical isolates (Machado et al., 2007; Sedighi et al., 2017). Fortini et al. (2011) has also reported dominance of bla (TEM) gene in Enterobacteriaceae isolated from Nigeria and only one strain was positive for the CTX-M gene. However, more prevalence of SHV gene has been reported in isolates from humans in Brazil (Dropa et al., 2009; Naim, Saeed & Malik, 2018). Prevalence of 96.7% ESBL has been previously reported in Klebsiella pneumonia isolated from commercial broiler slaughter plant in Shandong province of China (Wu et al., 2016). This is comparable with the results obtained in this study.
There seems to be a linkage between resistance to quinolones and the β-lactam antibiotics. It has been demonstrated that fluoroquinolone resistance can be mediated by co-transfer of the qnr determinant on ESBL-carrying plasmids (Wang et al., 2004) In some enterobacteriaceae, quinolone resistance is more frequent in ESBL positive stains than in ESBL-negative strains which may be due to combined effects of several mechanisms of resistance (Martinez-Martinez et al., 2002). Cremets et al. (2011) also noted that a CTX-M-15 producing isolate was highly resistant to fluoroquinolones and harboured mutations in the QRDR with two PMQR determinants.
Klebsiella pneumoniae strains had the highest level of resistance to antibiotics, which can be related to the presence of both ESBL and PMQR resistance genes. The most prevalent PMQR gene is oqxB with more abundance in Klebsiella pneumonia strains. (Yuan et al., 2012) and Szabó et al. (2018) also reported the presence of oqxAB gene in all human K. pneumoniae strains investigated. However, in a recent study on Klebsiella strains isolated from broiler, qnrB was the most dominant followed by qnrS and with low prevalence of qepA and qnrA (Wu et al., 2016).
In Morganella morganii, the prevalence of the three ESBL genes was relatively high with the TEM gene being the most prevalent. Al-Muhanna, Al-Muhanna & Alzuhari (2016) also showed that all Morganella morganii isolates tested for the ESBL had all the genes. Morganella morganii produces an inducible, chromosomally encoded AmpC β-lactamse, which can be implicated for its natural resistance to aminoopenicillins, amoxicillin-clavulanate, first and second generation cephalosporins with aztreonam, carbenicillin, and tazobactam being effective transient inactivators of some variants (Power et al., 2006). This is observed in this study, with most Morganella morganii being susceptible to aztreonam. Additionally, Morganella morganii is naturally resistant to tetracyclines, tigectycline, polymyxins and nitrofurantoin (Leclercq et al., 2013). However, intermediate resistance to tigercycline was observed in this study. Although high resistance to ciprofloxacin was observed in Morganella morganii in this study, they didn’t possess any of the PMQR genes investigated. The observed resistance may be due to an univestigated gene. Resistance of Morganella morganii to quinolones has been linked to qnrD, two new gyrB mutations (S463A, S464Y) and one parC mutation (S80I) (Nasri et al., 2014; Szabó et al., 2018).
All the Leclercia adecarboxylata species had the TEM gene, which may be responsible for their complete resistance to ceftazidime and cefotaxime. There was a moderate prevalence of SHV and CTX-M genes. Garcia-Fulgueiras et al. (2014) also reported the first presence of SHV and TEM genes in Leclercia adecarboxylata. The first case of CTX-M in Leclercia adecarboxylata was reported in a multi-drug resistant strain harbouring CTX-M and TEM gene (Shin et al., 2012). The prevalence of qep and oqxB gene was low in Leclercia adecarboxylata. In addition, three Morganella morganii and one Leclercia adecarboxylata had all three (SHV, TEM and CTX-M) genes; this increases the probability of these organisms developing resistance to quinolones.
Limitation of the study
The observed resistance to ciprofloxacin in Morganella morganii strains could be due to qnr D gene. However, the gene was not investigated in this study.
Conclusion
This study reports high prevalence of oqxB, TEM and SHV in poultry faecal dropping. To the best of our knowledge, this is the first study reporting occurrence of ESBL and PMQR in Morganella morganii and Leclercia adecarboxylata isolated from poultry.