CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data
- Published
- Accepted
- Received
- Academic Editor
- C. Titus Brown
- Subject Areas
- Bioinformatics
- Keywords
- Phylogenetics, Single nucleotide polymorphism, Mapping, Whole genome sequencing, Python, Validation
- Licence
- This is an open access article, free of all copyright, made available under the Creative Commons Public Domain Dedication. This work may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose.
- Cite this article
- 2015. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Computer Science 1:e20 https://doi.org/10.7717/peerj-cs.20
Abstract
The analysis of next-generation sequence (NGS) data is often a fragmented step-wise process. For example, multiple pieces of software are typically needed to map NGS reads, extract variant sites, and construct a DNA sequence matrix containing only single nucleotide polymorphisms (i.e., a SNP matrix) for a set of individuals. The management and chaining of these software pieces and their outputs can often be a cumbersome and difficult task. Here, we present CFSAN SNP Pipeline, which combines into a single package the mapping of NGS reads to a reference genome with Bowtie2, processing of those mapping (BAM) files using SAMtools, identification of variant sites using VarScan, and production of a SNP matrix using custom Python scripts. We also introduce a Python package (CFSAN SNP Mutator) that when given a reference genome will generate variants of known position against which we validate our pipeline. We created 1,000 simulated Salmonella enterica sp. enterica Serovar Agona genomes at 100× and 20× coverage, each containing 500 SNPs, 20 single-base insertions and 20 single-base deletions. For the 100× dataset, the CFSAN SNP Pipeline recovered 98.9% of the introduced SNPs and had a false positive rate of 1.04 × 10−6; for the 20× dataset 98.8% of SNPs were recovered and the false positive rate was 8.34 × 10−7. Based on these results, CFSAN SNP Pipeline is a robust and accurate tool that it is among the first to combine into a single executable the myriad steps required to produce a SNP matrix from NGS data. Such a tool is useful to those working in an applied setting (e.g., food safety traceback investigations) as well as for those interested in evolutionary questions.
Introduction
The application of whole genome sequence (WGS) data to problems in public health is currently undergoing rapid expansion. Within public health, next generation sequence (NGS) data provides superior discriminatory power compared to historical methods where it has, for example, been used to unequivocally trace the source of a number of Salmonella outbreaks (Allard et al., 2013; Lienau et al., 2011). Given the impact of regulatory decisions based on such data, there is a need for validated open-access and robust analysis tools that are relatively easy to use and support transparent and reproducible decision making.
The analysis of NGS data is a complex process within which multiple analysis steps are combined to produce a final result (e.g., a list of variant sites, a phylogeny, or a list of differentially expressed transcripts). Each of the steps often requires a different piece of software where the results of the previous step are the input for the next. Managing the files and processes of these steps can be inefficient and the results may not be easy to reproduce. This is problematic when the full analysis must be run across many data sets or run many times to assess sensitivity to parameter values.
A specific use of NGS data in public health is the determination of the relationship between samples potentially associated with a foodborne pathogen outbreak. This relationship can be determined from the phylogenetic analysis of a DNA sequence alignment containing only variable positions, which we refer to as a SNP matrix. Creation of a SNP matrix may include a number of steps, such as the mapping of reads to a reference genome, organization of the mapping files, identification of variant sites, calling of SNPs at each variant site, combining the SNPs into a SNP matrix, and summarizing the results of the process. For such work, a researcher would likely have to use at least three different pieces of software and track the results for each sample. Furthermore, one may be working with hundreds of samples, and these may accrue over time, further complicating the analyses.
Here, we describe CFSAN SNP Pipeline, which combines all the steps necessary to construct a reference-based SNP matrix from an NGS sample data set into a single, easy-to-use package. Having a single package facilitates the use of NGS data and SNP based analyses to identify differences among a set of closely related samples. The applications of such a matrix include inferring a phylogeny for systematic studies and determining within traceback investigations whether a clinical sample is significantly different from environmental/product samples.
Methods and Results
CFSAN SNP Pipeline architecture
The CFSAN SNP Pipeline is written in a combination of Bash and Python (it has been tested on Python 2.6 and 2.7). The code is designed to be straightforward to install (with pip) and has been tested on current versions of Red Hat, CentOS, and Ubuntu. When processing large datasets, the pipeline can take advantage of concurrent processing capabilities on high performance computing (HPC) systems or cloud-based clusters. The Bash scripts that manage the pipeline run seamlessly on a workstation or HPC with either Torque (Adaptive Computing, Provo, UT, USA) or Grid Engine (Univa Co., Lisle, Illinois, USA) job schedulers.
Scripts are provided to run the Python code from the command line. A configuration file allows customization of the pipeline, including changing the default parameter settings within each step of the pipeline. In situations where additional customization is desired, the code is not highly complex and can be modified as necessary. BioPython must be installed and there are also three executable software dependencies, Bowtie2 (Langmead & Salzberg, 2012), SAMtools (Li et al., 2009), and VarScan, (Koboldt et al., 2012). Configuration management of these dependencies is up to the user, as the dependencies are not supplied as part of the download. The CFSAN SNP Pipeline uses the versions of these tools found on the path. As new versions of the tools are released, the CFSAN SNP Pipeline is updated to include options associated with the new releases of the dependencies, which can be passed via the configuration file. The dependencies were chosen based on a combination of factors including being published in a peer-reviewed journal, performance metrics (e.g., Bowtie2; Langmead & Salzberg, 2012), and flexibility to handle haploid and diploid organisms (e.g., VarScan).
The CFSAN SNP Pipeline uses reference-based alignments to create a matrix of SNPs for a given set of samples. As a result, a reference in fasta format to which reads are mapped must be provided; suitable references include a high quality draft assembly or closed genome. For samples, sequence data must be in fastq format but can either be paired-end or single-read data. Three example datasets are provided with the release of the software (Table 1). The pipeline does not quality filter the fastq data so that must be done before running the program if desired (for issues surrounding quality filtering see e.g., Del Fabbro et al., 2013; Macmanes, 2014). However, certain steps within the pipeline filter results based on quality scores (e.g., variant detection with VarScan). Additionally, parameter settings can be altered to take advantage of the other software employed that can filter based on quality data (e.g., trimming with Bowtie2).
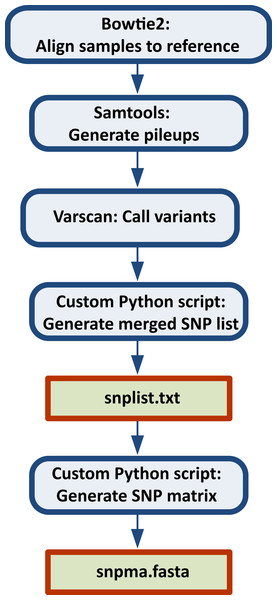
Once the reference sequence and sample data have been organized appropriately, the general steps in the CFSAN SNP Pipeline are as follows (Fig. 1):
Figure 1: Steps in the SNP Pipeline.
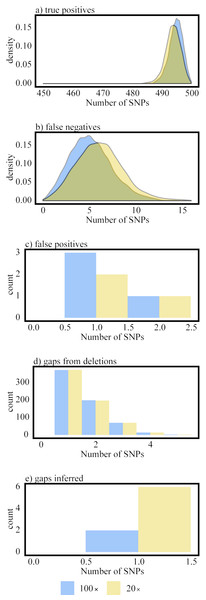
Rounded blue outlined boxes are analysis steps and squared red outlined boxes are files produced by the pipeline.Figure 2: Differences in performance between 100× and 20× datasets generated from the 1,000 mutated genomes.
(A) density plot of true positives; (B) density plot false negatives; (C) histogram of false positives; (D) histogram of gaps from indels; and (E) histogram of gaps inferred (i.e., gaps that were not originally mutated as an indel by CFSAN SNP Mutator).-
Create an indexed reference using Bowtie2.
-
Map sample reads to reference and create SAM files using Bowtie2.
-
Generate pileups from the SAM files using SAMtools.
-
Call variant sites (VCF file generation) from the pileup files using VarScan.
-
Using a custom script, generate a file (snplist.txt) containing all variant sites by aggregating across all VCF files.
-
Determine the base for each sample at each position in the snplist.txt file using a custom written consensus basecaller. We do not rely on VarScan to make the consensus call as it defaults to the nucleotide state of the reference when mapping quality falls below user-defined thresholds or the position is variant in one sample but missing in another.
-
Create a SNP matrix in fasta format for all samples where positions in the matrix are those found in snplist.txt.
All of these steps are run automatically, and only depend on the proper organization of the input files (see online documentation for further details) and identification of a suitable reference. Additionally, each of the above steps can either be run using individual shell scripts or the user can run a single shell script (run_ snp_pipeline.sh) that will carry out all steps in the pipeline. The addition of new samples is very straightforward, and result files from previous portions of the analysis that do not need to be re-generated are reused. This greatly reduces the computational time when adding new samples as the mapping and pileup steps are not re-executed.
CFSAN SNP Pipeline performance
To evaluate the performance of CFSAN SNP Pipeline, we developed a Python package (CFSAN SNP Mutator; https://github.com/CFSAN-Biostatistics/snp-mutator) to generate mutated genomes within which we know the positions where SNPs exist with respect to a reference. CFSAN SNP Mutator takes as input the reference sequence in fasta format, for example a closed bacterial reference genome, and based on user defined values will generate a number of replicate mutated genomes with a given number of single-base substitution and insertion/deletion polymorphisms. The output of CFSAN SNP Mutator is the mutated reference in fasta format and, if specified, a summary file of the differences from the reference. Each row of the summary file corresponds to one mutation with respect to the reference and contains four columns: (1) Replicate: identifier for the simulated genome; (2) Position: position in the reference that was mutated; (3) Original Base: the nucleotide state of the position in the reference and; (4) New Base: the mutation introduced, which will either be one of the four nucleotide states if a substitution was introduced, or “_deletion,” or two bases followed by “_insertion.”
Using CFSAN SNP Mutator, we generated 1,000 mutated genomes from the reference isolate (NCBI RefSeq Acces. NC_011149.1) that is part of the Agona test dataset for CFSAN SNP Pipeline (Table 1). Each mutated genome contained 500 substitution, 20 insertion, and 20 deletion polymorphisms. From these mutated genomes, we generated two datasets of simulated 250 bp paired end reads under the default Illumina MiSeq error profile using the program ART version ChocolateCherryCake (Huang et al., 2012). The two simulated fastq datasets had coverage depths of 20 and 100, which were then analyzed with the CFSAN SNP Pipeline (See Data S1 for instructions on how to recreate simulations analyzed here.). In the analysis of the reads with CFSAN SNP Pipeline we set the maximum fragment length for valid paired-end alignments to 547 via the –X Bowtie2 option. Given the parameters used for ART, this ensured that ∼99% of paired reads are considered concordant by Bowtie2; without adjusting this parameter half of our synthetic reads would have been considered ‘orphans’ by samtools and not included in the pileup (unless we had used the liberal—A flag with samtools).
One of the novel features of the CFSAN SNP Pipeline is the consensus snp caller. We analyzed all the mutated genomes simultaneously with one run of CFSAN SNP Pipeline. This will produce different results from running all of the mutated genomes individually because some mutated positions are shared among the 1,000 mutated genomes and some of these positions will not be detected individually by VarScan with the parameter settings we used (at least 8x coverage and 0.9 consensus frequency). However, the state for all samples at these shared mutated sites will be called using our consensus caller, and thus a difference at a site may be identified that was not detected by VarScan. This occurs in empirical datasets we work with where, for example, two samples are closely related and VarScan identifies variant positions within one sample that have not been detected in the other (due to a lower coverage (8x) or consensus frequency (0.9)). However, the actual nucleotide state found in the SNP matrix for such positions is called for both samples using our consensus caller.
The results from analyzing the genomes created by CFSAN SNP Mutator illustrate that the CFSAN SNP Pipeline has high true positive and true negative rates and low false positive and false negative rates (Table 2 and Fig. 2). The amount of coverage within the dataset did have some impact on the results but differences in performance were on the order of tenths of a percent. Not surprisingly, there is higher recovery rate of SNPs and a lower false negative rate within the 100× dataset compared to 20× . Looking closer at the causes for missed SNPs (false negatives), the vast majority of them within each dataset are due to a lack of consensus among reads where the frequency was below 0.9 (Table 3); as expected, failure to detect a SNP due to low coverage was higher in the 20× dataset. There were only 4 and 5 false positives detected in the 20× and 100× datasets, respectively (Fig. 2). The majority of false positives occurred at deletions that were a substitution in another sample (Table S1). For those samples with the false positive, the coverage at the deletion was only one. This leads to calling an incorrect nucleotide state rather than a gap because there is no threshold coverage in the consensus caller. This raises the caveat that incorrect states may be inferred with our consensus caller due to lack of a coverage threshold; future developments of the CFSAN SNP Pipeline will include such a threshold.
| 20× coverage | 100× coverage | |
|---|---|---|
| Coverage < 8 | 923 | 13 |
| Consensus frequency < 0.9 | 4,800 | 4,991 |
| Coverage < 8 & Consensus frequency < 0.9 | 63 | 2 |
| Coverage > 8 & Consensus frequency > 0.9 | 285 | 140 |
| Consensus = reference | 185 | 109 |
| Consensus ! = reference | 100 | 31 |
| Failed strand filter | 98 | 31 |
Although the CFSAN SNP Pipeline is designed to identify SNPs and not indels, we introduced the latter mutations to evaluate how they may impact the detection of variant positions within our pipeline. We found that the vast majority of the 20,000 deletions in each dataset were not identified as a variant unless that position was also mutated to a substitution in another sample (see Table S1 for exceptions). For example, in the 100× dataset 1,051 out of 1,053 gaps called by the CFSAN SNP Pipeline were deletions in one sample but those positions were also mutated to a substitution in another sample. There were also 6 and 2 gaps called by the CFSAN SNP Pipeline that were not introduced as a deletion into a genome by CFSAN SNP Mutator (Table 2). However, those positions were mutated in another sample and thus our consensus caller determined the state as a missing character because the allele frequency was below 0.6.
Of the 20,000 insertions introduced, none of them were called as a variant site in either dataset regardless of whether the position was also mutated in another sample.
Discussion
The CFSAN SNP Pipeline implements a robust and accurate methodology for constructing a matrix of SNPs for a given set of closely related sequences. The pipeline performs best when there is appreciable coverage at all sites, which was mostly the case in both the 20× and 100× datasets were investigated. This software was developed with the objective of creating high quality SNP matrices from WGS data of isolates derived from samples presumed to be involved with a single food-borne disease outbreak. The focus on closely related samples means that this code is not suited for the analysis of relatively distantly related organisms (e.g., with regards to bacteria, greater than a few hundred SNP differences), where there is likely not a single appropriate reference sequence (see Bertels et al., 2014; Pightling, Petronella & Pagotto, 2014 for issues surrounding the use of a distant reference). Furthermore, although a number of reference free, easy to use packages have been developed to construct SNP matrices for phylogenetic analyses (e.g., Gardner & Hall, 2013; Schwartz et al., 2013), those reference free approaches suffer from an increased false-discovery rate and are not as conservative as reference based approaches (Pettengill et al., 2014).
The CFSAN SNP Pipeline documentation provides examples of using the code as well as three test data sets (Table 3). We also developed a package to generate mutated genomes within which the variants are known. These test and simulated datasets serve as unit tests that allows for the verification that changes to the code have not changed the results produced. The simulated data used here for validation, that can be reproduced based on information in Data S1, also provides data that others can use to compare the results obtained from their analysis methodology. Such datasets, in which the variants are known, appear to be lacking within the arena of variant callers like CFSAN SNP Pipeline.
The SNP matrix produced with our pipeline can be used for a number of different purposes. Our particular focus is using such a matrix to construct a phylogeny to resolve outbreaks of foodborne pathogens, and, thus the CFSAN SNP Pipeline is a particularly useful and necessary tool for those within public health. We are only aware of one other package that has bundled together many of the steps for creating a SNP matrix using a reference-based approach (Lee et al., 2014), but that method assumes that user has variants already in VCF files. The CFSAN SNP Pipeline represents one of the first complete tools for constructing a SNP matrix from raw reads and a reference.
Availability and requirements
Project name: CFSAN SNP Pipeline
Source code: https://github.com/CFSAN-Biostatistics/snp-pipeline
Documentation: http://snp-pipeline.rtfd.org
PyPI package: https://pypi.python.org/pypi/snp-pipeline
Operating system: Linux (Red Hat, CentOS, and Ubuntu)
Programming language: Bash and Python
Other requirements: Java v1.3.1 or higher, Bowtie2, sra-toolkit, SAMtools, VarScan, BioPython. (Note: use of version 2.3.6 of VarScan is not recommended due to occasional omission of the header information in the output .vcf files that cause problems with the CFSAN SNP Pipeline. Use of version 2.3.9 is known to work properly).
License: As a work of the United Stated Government, CFSAN SNP Pipeline is not subject to copyright protection and will remain freely available.
Any restrictions to use by non-academics: None