Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth
- Published
- Accepted
- Received
- Academic Editor
- Pedro Silva
- Subject Areas
- Agricultural Science, Microbiology
- Keywords
- Biofertilizer, Paenibacillus, Nitrogenase, Plant growth promotion, Plant pathogens, IAA
- Copyright
- © 2019 Liu et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth. PeerJ 7:e7445 https://doi.org/10.7717/peerj.7445
Abstract
Paenibacillus is a large genus of Gram-positive, facultative anaerobic, endospore-forming bacteria. The genus Paenibacillus currently comprises more than 150 named species, approximately 20 of which have nitrogen-fixation ability. The N2-fixing Paenibacillus strains have potential uses as a bacterial fertilizer in agriculture. In this study, 179 bacterial strains were isolated by using nitrogen-free medium after heating at 85 °C for 10 min from 69 soil samples collected from different plant rhizospheres in different areas. Of the 179 bacterial strains, 25 Paenibacillus strains had nifH gene encoding Fe protein of nitrogenase and showed nitrogenase activities. Of the 25 N2-fixing Paenibacillus strains, 22 strains produced indole-3-acetic acid (IAA). 21 strains out of the 25 N2-fixing Paenibacillus strains inhibited at least one of the 6 plant pathogens Rhizoctonia cerealis, Fusarium graminearum, Gibberella zeae, Fusarium solani, Colletotrichum gossypii and Alternaria longipes. 18 strains inhibited 5 plant pathogens and Paenibacillus sp. SZ-13b could inhibit the growth of all of the 6 plant pathogens. According to the nitrogenase activities, antibacterial capacities and IAA production, we chose eight strains to inoculate wheat, cucumber and tomato. Our results showed that the 5 strains Paenibacillus sp. JS-4, Paenibacillus sp. SZ-10, Paenibacillus sp. SZ-14, Paenibacillus sp. BJ-4 and Paenibacillus sp. SZ-15 significantly promoted plant growth and enhanced the dry weight of plants. Hence, the five strains have the greater potential to be used as good candidates for biofertilizer to facilitate sustainable development of agriculture.
Introduction
Nitrogen is an essential element to affect the yields of crops by influencing leaf area development and photosynthetic efficiency (Fang et al., 2018). The application of chemical nitrogen fertilizer can improve soil fertility and thus agricultural production. High rates of nitrogen fertilizer might boost yields, but can reduce the quality of agricultural products. However, approximately 100 Tg chemical nitrogen is applied in agricultural products every year, while only 17 Tg nitrogen is accounted for in crops (Erisman et al., 2008). Excessive use of chemical fertilizer has resulted in seriously negative impacts, such as soil hardening and acidification, increased greenhouse gas (N2O) emissions and enhanced nitrogen deposition (Jiao et al., 2018; Reay et al., 2012).
The Paenibacillus genus was first reclassified as a separate genus on the basis of the 16S rRNA gene sequences by Ash, Priest & Collins (1993). Since its creation, the Paenibacillus genus embody more than 100 validly named species. Approximately 20 members of the Paenibacillus genus had been reported to have the capacity of fixing nitrogen, such as: Paenibacillus polymyxa, Paenibacillus macerans, Paenibacillus azotofixans, Paenibacillus sabinae, Paenibacillus sonchi, Paenibacillus forsythia, Paenibacillus sophorae, Paenibacillus taohuashanense and Paenibacillus beijingensis (Grau & Wilson, 1962; Hong et al., 2009; Jin, Lv & Chen, 2011; Ma et al., 2007; Ma & Chen, 2008; Seldin, Van Elsas & Penido, 1984; Wang et al., 2014; Witz, Detroy & Wilson, 1967; Xie et al., 2012). Paenibacillus is a group of Gram-positive, aerobic or facultative anaerobic, rod-shaped, endospore-forming bacteria. The widely distributed Paenibacillus bacteria could tolerate extreme environments and interact with a variety of plants (Navarro-noya et al., 2012). Currently, some Paenibacillus strains play a great role in agriculture and industry (Seldin, 2011).
Plant rhizosphere is a habitat of functional microorganisms, which encompasses a complex and dynamic zone of interactions between networks of organisms and their plant hosts (Garcia & Kao-Kniffin, 2018; Zhalnina et al., 2018). A large amount of strains isolated from plant rhizospheres are able to directly or indirectly promote plant growth, development and evolution, which are termed as plant growth-promoting rhizobacteria (PGPR) (Mohamed et al., 2019). PGPR can stimulate plant growth by a diversity of mechanisms including fixing nitrogen from atmosphere, solubilizing phosphorus, synthesizing siderophore, producing antimicrobial substances (antibiotics, bacteriocins and small peptides) and plant hormones such as indole, cytokinins or gibberellins (Graham et al., 2000; Neilands, 1993). Given these advantages, PGPR are widely used in sustainable agriculture to promote plant growth and control fungal pathogens (Verma et al., 2018). Some of Paenibacillus species can influence plant growth by one or more of mechanisms mentioned above (Li et al., 2017; Weselowski et al., 2016; Xie et al., 2016). Nowadays, with the rapid growth of population, most regions have increased the cereals production by the overuse of fertilizers, which not only accounts for a larger percentage of farmers’ expenses but also increase risks of negative effect on environment (Curatti & Rubio, 2014; Ivleva et al., 2016; Tayefeh et al., 2018). It is the best choice to select the environmentally friendly Paenibacillus strains to substitute for chemical fertilizer due to its broad host range and its ability to secrete plant growth-enhancing substances and produce different kinds of antimicrobial substances (Cho et al., 2007; Da Mota, Gomes & Seldin, 2008; Fortes et al., 2008; Li et al., 2007; Timmusk et al., 2009).
The Paenibacillus strains have the potential to increase agricultural productivity, including weight of crops and root growth. The main purpose of this research was to isolate and identify Paenibacillus strains, to study the effect of these isolates on plant growth, and then to select the potential bacterial strains to be used in sustainable development of agricultural production.
Materials & Methods
Sample collection, isolation procedures and culture conditions
Sixty-nine soil samples were collected from various plant rhizospheres in different areas of China, which were described in Table 1 in detail. The soil samples were diluted gradiently by 0.9% saline solution (up to 10−5) and then screened on nitrogen-free medium after heating at 85 °C for 10 min. Three replicates per dilution were made. The nitrogen-free medium contained 20 g sucrose, 0.1 g K2HPO4, 0.4 g KH2PO4, 0.2 g MgSO4 7H2O, 0.01 g NaCl, 0.01 g FeCl3, 0.002 g Na2MoO4 and 1.2–1.4 g agar per litre of water. Single colony for each possible species was selected after cultivation for 3–5 days at 30 °C. To reduce the influence of nitrogen from the soils and purify the strains, the isolates were transferred to the fresh nitrogen-free medium. The strains isolated in this study and their sources were listed in Table 1. All isolates are stored in our lab, and 16S rRNA sequences are available in database of GenBank.
| Isolates | Cell morphology | Colony morphology | Nitrogenase activitya | GenBank accession number | Origin and location |
|---|---|---|---|---|---|
| Paenibacillus sp. BJ-2 | Rods | Moist, milky | 1,085.61 ± 75.64ghi | MF967282 | Jujube, mountain in Huairou, Beijing 40°32′N, 116°62′E |
| Paenibacillus sp. SZ-1a | Rods | Moist, milky | 118.65 ± 3.97k | MF967283 | Maize, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-1b | Rods | Moist, milky | 1,1868.65 ± 1740.55a | MF967284 | Maize, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. BJ-4 | Rods | Dry, milky | 1,296.94 ± 439.17g | MF967285 | Apple, orchard in Shunyi, Beijing 40°13′N, 116°65′E |
| Paenibacillus sp. BJ-5 | Rods | Dry, white | 468.63 ± 42.20hijk | MF967286 | Persimmon, mountain in Shunyi, Beijing 40°13′N, 116°65′E |
| Paenibacillus sp. SZ-8 | Rods | Moist, milky | 1,131.54 ± 15.92gh | MF967287 | Maize, field in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. BJ-7 | Rods | Moist, milky | 314.60 ± 19.18jk | MF967288 | Wheat, farmland in Miyun, Beijing 40°37′N, 116°85′E |
| Paenibacillus sp. SZ-10 | Short rods | Moist, milky | 371.28 ± 7.67ijk | MF967289 | Maize, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-11 | Rods | Moist, milky | 857.47 ± 114.89ghij | MF967290 | Pepper, herbary in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-13a | Rods | Dry, milky | 9,731.36 ± 259.71b | MF967291 | Medicinal plant, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-13b | Rods | Dry, milky | 3,131.89 ± 100.61e | MF967292 | Medicinal plant, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-15 | Rods | Moist, milky | 1,316.19 ± 36.64g | MF967293 | Wheat, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. SZ-16 | Rods | Moist, milky | 444.73 ± 119.11hijk | MF967294 | Spinach, herbary in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. BJ-6 | Short rods | Dry, milky | 176.7 ± 29.43jk | MF967295 | Bamboo, mountain in Huairou, Beijing 40°32′N, 116°62′E |
| Paenibacillus sp. AH-1 | Short rods | Moist, milky | 192.43 ± 73.08jk | MF967296 | Grape, orchard in Hefei, Anhui 31°86′N, 117°27′E |
| Paenibacillus sp. SZ-14 | Rods | Moist, milky | 331.95 ± 22.73jk | MF967297 | Rice, farmland in Changping, Beijing 40°22′N, 116°20′E |
| Paenibacillus sp. YN-3 | Short rods | Moist, white | 3,201.92 ± 104.96e | MF967298 | Sugarcane, farmland in Pu’er, Yunnan 23°07′N, 110°03′E |
| Paenibacillus sp. AH-3 | Short rods | Moist, white | 57.23 ± 14.44k | MF967299 | Arbor, natural forest in Wuhu, Anhui 31°95′N, 118°73′E |
| Paenibacillus sp. AH-4 | Short rods | Moist, white | 6,514.37 ± 997.12c | MF967300 | Arbor, natural forest in Hefei, Anhui 31°95′N, 118°73′E |
| Paenibacillus sp. YB-3 | Rods | Moist, milky | 733.92 ± 49.28ghijk | MF967301 | Fruit, mountain in Yibin, Sichuan 28°77′N, 104°62′E |
| Paenibacillus sp. WF-6 | Rods | Moist, milky | 2,081.30 ± 340.66f | MF967302 | Wheat, field in Weifang, Shandong 36°62′N, 119°10′E |
| Paenibacillus sp. JS-4 | Rods | Moist, milky | 6,843.56 ± 365.69c | MF967303 | Reed, countryside in Suzhou, Jiangsu 31°32′N, 120°62′E |
| Paenibacillus sp. HN-1 | Short rods | Moist, milky | 4,476.80 ± 306.64d | MF967304 | Rice, farmland in Xiangtan, Hunan 27°52′N, 112°53′E |
| Paenibacillus sp. CD-4a | Rods | Moist, milky | 272.67 ± 14.24jk | MF967305 | Rape, field in Chengdu, Sichuan 30°67′N, 104°07′E |
| Paenibacillus sp. CD-4b | Short rods | Moist, milky | 5,174.69 ± 478.7d | MF967306 | Fruit, mountain in Chengdu, Sichuan 30°67′N, 104°07′E |
Notes:
Results are means ± SE of 3 independent biological replicates. Different letters are significantly different from each other according to the least significant differences (LSD) test (P < 0.05).
Amplification, cloning and sequencing of nifH gene
PCR amplification of nifH gene was carried out using the following primers: forward 5′-GGCTGCGATCC(CGA)AAGGCCGATC(CGA)ACCCG-3′ and reverse 5′-CTG(GCA)GCCTTGTTTCGCGGAT(CG)GGCATGGC-3′ as described by Ding et al., (2005). The nifH gene fragments were purified using TIANgel Midi Purification Kit (Tiangen Biotech Co., LTD. Cat. #DP210, Beijing, China) and ligated to vector pGEM-T (Promega Co., Cat. #R6881; Madison, WI, USA) at 16 °C overnight. Recombinant plasmids were transformed into Escherichia coli JM109 and transformants were selected by blue/white screening procedure. Plasmids containing nifH gene were extracted and purified. Purified plasmids were then sequenced using the M13F and M13R primers by Shanghai Majorbio Bio-pharm Technology Co., LTD, Shanghai, China.
Morphological characterization of strains
For observation of colony morphology, the bacterial strains were spread on Luria-Bertani (LB) agar. After incubation at 37 °C overnight, single colony was observed. Cell morphology was viewed by optical microscopy (Olympus, CX22LED, Tokyo, Japan).
Sequence analysis and construction of the phylogenetic trees
All strains were cultured in LB broth medium overnight. After collection of bacteria by centrifugation, genomic DNA of isolates was extracted and purified using the TIANamp Bacteria DNA Kit (Tiangen Biotech Co., LTD. Cat. #DP302) according to the manufacturer’s instructions. The amplication of 16S rRNA genes was performed with the universal primers: 27F (5′-AGAGTTTGATC(AC)TGGCTCAG-3′) and 1492R (5′-CGG(CT)TACCTTGTTACGACTT-3′) as described by Khan et al. (2014). Then the 16S rRNA gene fragments were ligated into vector pGEM-T (Promega Co., Cat. #R6881) and sequenced by Shanghai Majorbio Bio-pharm Technology Co., LTD. The sequences of 16S rRNA gene were submitted to nucleotide database of GenBank and the accession numbers were displayed in Table 1. And the sequences were aligned with BLAST software from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The phylogenetic tree was constructed from evolutionary distance matrices using the neighbor-joining method with MEGA6 software package (Tamura et al., 2013). Bootstrap analysis was performed with 1,000 cycles, and only bootstrap values greater than 50% were shown at the branch points.
Nitrogenase activity assay
For determination of the nitrogenase activity, strains were grown in 20 mL of LB broth medium in 50 mL flasks shaken at 200 rpm overnight at 37 °C. The cultures were collected by centrifugation, precipitations were washed three times with sterilized water and then resuspended in nitrogen-limited medium (per liter: 26.3 g Na2HPO412H2O, 3.4 g KH2PO4, 26 mg CaCl2 2H2O, 30 mg MgSO4, 0.3 mg MnSO4, 36 mg ferric citrate, 7.6 mg Na2MoO4 2H2O, 10 µg p-aminobenzoic acid, 10 µg biotin, 0.4% (w/v) glucose and 0.03% (w/v) glutamic acid). The nitrogenase activity was determined using the acetylene reduction assay and expressed as nmol C2H4 mg−1 protein h−1 (Wang et al., 2013; Wang et al., 2018).
Assessment of antagonistic activity against plant pathogens
The assessment of the Paenibacillus strains isolated from the rhizospheres for antagonism against 6 plant pathogens including Rhizoctonia cerealis (ACCC 37393), Fusarium graminearum (ACCC 36249), Gibberella zeae (CGMCC 3.2873), Fusarium solani (CGMCC 3.17848), Colletotrichum gossypii (CGMCC 3.1859) and Alternaria longipes (CGMCC 3.2875), was performed in agar plate assay using potato dextrose agar (PDA). The fungal pathogens were inoculated in the center of the agar plate, and the Paenibacillus strains were placed at a distance of 3.5 cm from the center of the plate. After 3–7 days of incubation at 30 °C, the plates were examined and measured for fungal pathogens growth inhibited zones around the Paenibacillus strains. All tests were carried out in three duplicates.
Measurement of indole-3-acetic acid (IAA) production
The ability of producing IAA was assessed by colorimetric analysis. For the measurement of IAA production, the tested strains were grown in 20 mL King B broth medium (per liter: peptone, 20 g; K2HPO4, 1.15 g; MgSO4 7H2O, 1.5 g; glycerol, 10 g) supplemented with 100 µg mL−1 Trp (IAA precursor). The non-cultured medium was used as the negative control and Azospirillum brasilense SP7 was selected as the positive control. The culture supernatants were obtained by centrifuging at 12,000 rpm for 10 min. The test strains were measured by colorimetric assay according to the method described by Glickmann & Dessaux (1995). Briefly, two mL Salkowski reagent containing 4.5 g/L FeCl3 in 10.8 M H2SO4 was mixed with one mL supernatant. Then, the mixture was stired evenly and left in the darkness for 30 min at room temperature. The production of IAA was measured using spectrophotometer (Shimadzu UVmini-1240; Kyoto, Japan) at 530 nm. Each treatment had three biological replicates.
Evaluation of plant growth-promoting effect
The tested strains were evaluated for their potential to promote plant growth on wheat cultivar Jimai 22 (Shandong Runfeng Seed Industry Co., Ltd., Shandong Sheng, China), cucumber Zhongnong 8 (Beijing Shengfeng Garden Agricultural Technology Co., Ltd., Beijing, China) and tomato Jiafen 15 (Tianjin Xingke Seed Co., Ltd., Tianjin, China) seedlings in the greenhouse of China Agricultural University, Beijing, China. The lengths and dry weights of three plants inoculated with strains were determined by the procedure described by Li et al. (2017).
For preparing the bacterial cultures, each isolate was grown 150 mL LB broth medium for 24 h at 30 °C. After incubation, the cells were harvested by centrifugation at 6,000 rpm for 5 min at room temperature. The cell pellet was washed with sterile water and then adjusted to 108 cells mL−1 with 0.9% saline solution.
Wheat, cucumber and tomato seeds were sterilized with 10% sodium hypochlorite for 10 min and washed with sterilized water three times. Then the seeds germinated on sterile wet filter in Petri dishes in the dark at 25 °C for 5–7 days. After germination, seedlings were soaked in bacterial suspensions (108 cells mL−1) for 15 min. Then three seedlings of different plants were transplanted into 12-cm-diam pots containing in the medium of turfy soil (Beijing Jixiang Feiyun Garden Engineering Co., Ltd. Cat. #101G): vermiculite (Beijing Jixiang Feiyun Garden Engineering Co., Ltd. Cat. #GM010108) of 1:1, and grown in the greenhouse (16 h day/8 h night and 22 °C/10 °C day/night temperature). Each treatment had three pots. Two weeks later, each of the seedlings was watered with 15 mL bacterial suspensions (108 cells mL−1) again. The un-inoculated seedlings were used as negative controls, while the un-inoculated seedlings watered with nitrogen fertilizer (83 mg N kg−1 soil) were set as positive controls (Li et al., 2019). After five-week growth, the plants were harvested and the roots were washed carefully with running water to remove the adherent soil. The lengths of the shoot and root and dry weights of the shoot and root were recorded and statistically analyzed, respectively.
Statistical analysis
Each treatment had three replicates. Statistical analysis was performed using SPSS 20.0 (SPSS, Chicago, IL, USA). Means of different treatments were compared using the least significant difference (LSD) at 0.05 level of probability.
Ethics approval and consent to participate
Not applicable.
Results
The nifH gene analysis and nitrogenase activity assay
Nitrogenase is comprised of two component proteins: Fe protein and MoFe protein (Mus et al., 2018). The Fe protein is encoded by nifH gene, and MoFe protein is encoded by nifD and nifK genes. The conserved nifH gene has been exploited to screen the genetic potential for nitrogen-fixing bacteria in the environment (Ding et al., 2005; Mehta, Butterfield & Baross, 2003).
In this study, 179 strains were isolated by using nitrogen-free medium after heating at 85 °C for 10 min from 69 soil samples collected from different plant rhizospheres in different areas. PCR amplification of nifH gene (encoding Fe protein of nitrogenase) with universal primers was conducted using genomic DNA extracted from above bacteria. The results showed that a nifH gene fragment of 323 nucleotides was detected in 25 isolates (Table 1). The PCR-amplified nifH gene fragments from 25 isolates were sequenced and their predicted amino acid sequences of NifH were aligned with the NifH sequences from other diazotrophs. The results showed that all of them except for Paenibacillus sp. HN-1 shared 84%–99% NifH sequence identity with other Paenibacillus strains. The sequencing result of Paenibacillus sp. HN-1 nifH fragment displayed double peaks, which indicated that there were multiple nifH genes in its genome.
As displayed in Table 1, all of the 25 strains with nifH genes had nitrogenase activities with variation from 57.23 to 11,868.65 nmol C2H4 mg−1 protein h−1. Paenibacillus sp. SZ-1b presented the highest nitrogenase activity (11868.65 nmol C2H4 mg−1 protein h−1). Paenibacillus sp. SZ-13a, Paenibacillus sp. SZ-13b, Paenibacillus sp. YN-3, Paenibacillus sp. AH-4, Paenibacillus sp. JS-4 and Paenibacillus sp. CD-4b had higher nitrogenase activities (>,3000 nmol C2H4 mg−1 protein h−1). The nitrogenase activity, cell morphology, colony morphology, GenBank accession number and origin/location were listed in Table 1.
Sequencing and phylogeny of 16S rRNA
The 16S rRNA gene sequence is named as the evolution clock of bacterial phylogeny because of high conservation and slow evolution, which is widely used in identification of bacteria (Roller et al., 1994; Vandamme et al., 1996). The 16S rRNA gene sequences of the 25 strains were compared with the datebase reserved in GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The alignment results indicated all of the isolates were Paenibacillus. The GenBank accession numbers of them after the bacterial names were shown in Table 1.
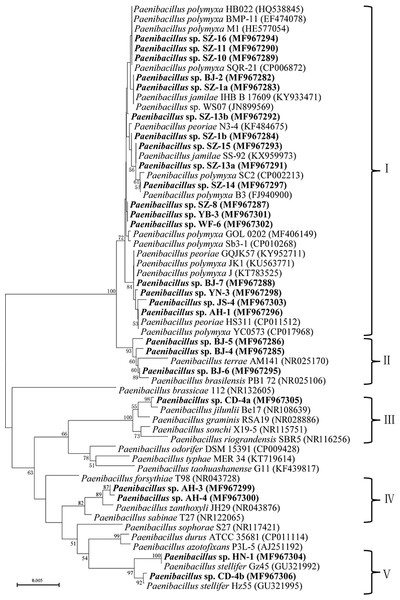
A phylogenetic tree was constructed based on 16S rRNA sequence, which branched into five clusters on the basis of the distance data. The cluster I totally including 17 isolates formed a larger cluster with P. polymyxa, Paenibacillus jamilae and Paenibacillus peoriae. Among the 17 isolates, six isolates exhibited 99.2%–99.6% 16S rRNA sequence similarities with P. polymyxa. 7 isolates had the highest similarities with P. jamilae, and four isolates showed particularly high homologies with P. peoriae (>99.5%). The cluster II contained three isolates, which displayed the highest similarity with Paenibacillus brasilensis, ranging from 99% to 99.2%. The cluster III only included Paenibacillus sp. CD-4a, which had highest 16S rRNA sequence similarity with Paenibacillus jilunlli (99.6%). The cluster IV which consisted of two strains clustered with Paenibacillus zanthoxyli showing 99.3% to 99.6% 16S rRNA sequence similarities with P. zanthoxyli. The cluster V covering two isolates formed a monophyletic cluster with Paenibacillus stellifer bacteria, and their 16S rRNA sequences similarities with P. stellifer were above 99%.
Antibacterial capacity determination
In the study, all 25 Paenibacillus strains were tested against six plant pathogens. The results (Table 2) showed that 21 bacteria presented antibiosis, inhibiting at least one of the 6 indicator phytopathogens. Out of them, 18 bacteria could inhibit five plant pathogens (R. cerealis, F. graminearum, G. zeae, C. gossypii and A. longipes). Furthermore, Paenibacillus sp. SZ-13b exhibited an extremely good antibiotic activity, which was able to inhibit the growth of all indicator phytopathogens. The growth of F. graminearum was strongly inhibited, showing the average inhibition zones larger than 25 mm. While the growth of F. solani was weakly inhibited, which was only inhibited by two strains (Paenibacillus sp. SZ-13b and Paenibacillus sp. BJ-6) with the inhibition zones around 5 and 15 mm. In addition, Paenibacillus sp. AH-3, Paenibacillus sp. HN-1, Paenibacillus sp. CD-4a and Paenibacillus sp. CD-4b could not exhibit any antibiotic effect on six indicator fungi.
| Strains | R. cer | F. gra | G. zeae | F. sol | C. gos | A. lon |
|---|---|---|---|---|---|---|
| Paenibacillus sp. BJ-2 | + + | + + | + + | – | + + + | + |
| Paenibacillus sp. SZ-1a | + + | + + | + + | – | + + | + + |
| Paenibacillus sp. SZ-1b | ++ + | ++ + | + + | – | + + | ++ + |
| Paenibacillus sp. BJ-4 | + + | ++ + | + + | – | + + | ++ + |
| Paenibacillus sp. BJ-5 | – | ++ + | + | – | + + | + + |
| Paenibacillus sp. SZ-8 | + + | ++ + | + + | – | ++ + | + + |
| Paenibacillus sp. BJ-7 | + + | ++ + | ++ + | – | + + | + |
| Paenibacillus sp. SZ-10 | + + | ++ + | ++ + | – | + + | + + |
| Paenibacillus sp. SZ-11 | + + | + + | + + | – | + + | + + |
| Paenibacillus sp. SZ-13a | ++ + | + + | + + | – | + + | + + |
| Paenibacillus sp. SZ-13b | + + | + + | + + | + | + + | |
| Paenibacillus sp. SZ-15 | ++ + | + + | ++ + | – | ++ + | + + |
| Paenibacillus sp. SZ-16 | + + | + + | + + | – | + + | + + |
| Paenibacillus sp. BJ-6 | + + | + + | – | + | + + | – |
| Paenibacillus sp. AH-1 | ++ + | ++ + | ++ + | – | + + | + + |
| Paenibacillus sp. SZ-14 | + + | ++ + | + + | – | ++ + | + + |
| Paenibacillus sp. YN-3 | + + | + + | + + | – | + + | + + |
| Paenibacillus sp. AH-3 | – | – | – | – | – | – |
| Paenibacillus sp. AH-4 | + | – | – | – | – | – |
| Paenibacillus sp. YB-3 | + + | ++ + | + + | – | + + | ++ + |
| Paenibacillus sp. WF-6 | + + | + + | + + | – | + + | + + |
| Paenibacillus sp. JS-4 | + + | ++ + | + + | – | ++ + | ++ + |
| Paenibacillus sp. HN-1 | – | – | – | – | – | – |
| Paenibacillus sp. CD-4a | – | – | – | – | – | – |
| Paenibacillus sp. CD-4b | – | – | – | – | – | – |
Notes:
- R. cer
-
R. cerealis
- F. gra
-
F. graminearum
- G. zeae
-
G. zeae
- F. sol
-
F. solani
- C. gos
-
C. gossypii
- A. lon
-
A. longipes
- (–)
-
no inhibition
- (+)
-
inhibition zone diameters from 5 to 15 mm
- (++)
-
inhibition zone diameters from 15 to 25 mm
- (+++)
-
inhibition zone diameters from 25 to 35 mm
In general, out of the 25 tested strains, 80% strains presented antimicrobial activity against plant pathogens, with average inhibition zones varying from 15 to 35 mm. Combination with their phylogeny of 16S rRNA, the isolates with inhibition flocked together, which were particularly close to P. polymyxa and its highly close species (Fig. 1).
Figure 1: Neighbour-joining phylogenetic tree based on 16S rRNA sequence showing the position of isolated strains with other closely related strains of the genus Paenibacillus in GenBank.
The tree was structured using neighbor joining method, with the bootstrap percentage values obtained from 1,000 cycles. Only bootstrap values greater than 50% are shown at the branching points. Bar, 0.005 substitutions per nucleotide position. Isolated strains in this study are underlined with the bold letters.Assessment of IAA production and plant growth promoting traits
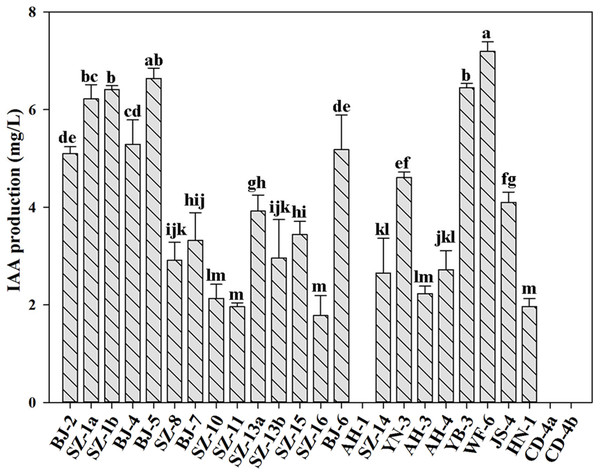
IAA is an essential plant hormone regulating the growth and development of plants. In this study, we determined the ability of producing IAA for all strains. Figure 2 showed that besides Paenibacillus sp. AH-1, Paenibacillus sp. CD-4a and Paenibacillus sp. CD-4b, the rest of tested strains were capable of producing IAA. Out of them, Paenibacillus sp. WF-6 produced the highest yield of IAA (7.19 mg L−1). In addition, the other nine bacteria (Paenibacillus sp. BJ-2, Paenibacillus sp. SZ-1a, Paenibacillus sp. SZ-1b, Paenibacillus sp. BJ-4, Paenibacillus sp. BJ-5, Paenibacillus sp. BJ-6, Paenibacillus sp. YN-3, Paenibacillus sp. YB-3, Paenibacillus sp. JS-4) could yield relatively high amount of IAA (>4 mg L−1).
Figure 2: Qualitative analysis of IAA production by isolated strains.
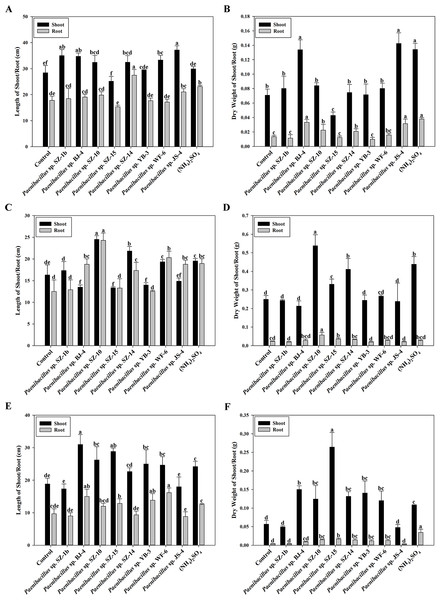
Data are means ± SE of threeindependent biological replicates. Bearing different alphabets are significantly different from each other according to the LSD test (p < 0.05).According to above results of nitrogenase activities, antibacterial capacities and IAA production, we chose eight strains (Paenibacillus sp. SZ-1b, Paenibacillus sp. BJ-4, Paenibacillus sp. SZ-10, Paenibacillus sp. SZ-13b, Paenibacillus sp. SZ-14, Paenibacillus sp. YB-3, Paenibacillus sp. WF-6, Paenibacillus sp. JS-4) to assess their capabilities of promoting growth of plants (wheat, cucumber and tomato). Inoculation of plants with some Paenibacillus isolates appeared to promote plant growth including plant height and dry weight (Figs. 3 and 4). As shown in Fig. 4A, wheat seedlings inoculated with Paenibacillus sp. JS-4 led to a maximum increase (30.9%) in shoot length, followed by Paenibacillus sp. SZ-1b (23.3%) and Paenibacillus sp. BJ-4 (22.3%). While inoculation with Paenibacillus sp. SZ-14 yielded a maximum increase (54.2%) in root length, followed by Paenibacillus sp. JS-4 (18.2%). Inoculation of wheat plants with Paenibacillus sp. JS-4 showed a greatly significant increase in shoot and root dry weights. Besides, Paenibacillus sp. BJ-4 and Paenibacillus sp. SZ-10 had higher dry weights of shoot and root as compared to the controls (Fig. 4B). The effects of these two bacteria on wheat seedlings were equal to the positive control with chemical nitrogen fertilizer. In Fig. 4C, cucumber seedlings inoculated with Paenibacillus sp. SZ-10 resulted in the highest heights both in shoot (50.0%) and in root (94.4%), followed by Paenibacillus sp. SZ-14 (33.7% and 38.7%, respectively) and Paenibacillus sp. WF-6 (18.4% and 62.4%, respectively). In addition, inoculation with Paenibacillus sp. SZ-10 presented the highest increase in dry weights of shoot and root of eight selected isolates, which showed more significant effect on cucumber seedlings than the positive control. Also, inoculation with Paenibacillus sp. SZ-14 had the second highest increase in total dry weight (Fig. 4D), which was the same as the positive control with chemical nitrogen fertilizer. Overall, Paenibacillus sp. SZ-10 showed significant growth-promoting effects on the cucumber plants. As shown in Figs. 4E and 4F, most isolates could promote growth of tomato. Out of them, inoculation with Paenibacillus sp. BJ-4 presented to enhance development of tomato length, both in shoot (64.6%) and in root (55.2%) (Fig. 4E). Inoculation with Paenibacillus sp. SZ-15 displayed maximum increases in shoot and root dry weights (Fig. 4F), which showed more promotive effect on shoot dry weight of tomato than the positive control.
Figure 3: Plant growth promotion by some Paenibacillus strains.
(A) Wheat seedlings inoculated with Paenibacillus sp. JS-4; (B) Cucumber seedlings inoculated with Paenibacillus sp. SZ-10; (C) Tomato seedlings inoculated with Paenibacillus sp. SZ-15.Figure 4: Effects of eight selected strains inoculation on shoot and root length of wheat (A), dry weight of wheat (B), on shoot and root length of cucumber (C), dry weight of cucumber (D), on shoot and root length of tomato (E), dry weight of tomato (F).
Control: un-inoculated seedlings. Date represent the means ± SE of 3 independent biological replicates. In the root group or shoot group, bearing different alphabets are significantly different from each other according to the LSD test (p < 0.05).Discussion
Paenibacillus species are ubiquitous in nature, and they are capable to form resistant endospores to allow them surviving in a wide range of environmental variables and to enhance plant growth by several mechanisms (Bloemberg & Lugtenberg, 2001). In this study, 179 bacterial strains were isolated by their growth on nitrogen-free medium from plant rhizospheres all over China. 16S rRNA sequence analysis showed that 25 of 179 bacteria belonged to Paenibacillus genus.
We revealed that 25 Paenibacillus strains had the nifH gene encoding the Fe protein of Mo-nitrogenase. Also, the 25 Paenibacillus strains exhibited nitrogenase activities. These results demonstrated that the 25 N2-fixing Paenibacillus strains could provide nitrogen for plants. Phylogenetic analysis showed that the 25 N2-fixing Paenibacillus strains were divided into five clusters. 20 of the 25 N2-fixing Paenibacillus strains were in cluster I and cluster II that were closely related to P. polymyxa, P. jamilae, P. peoriae, and P. brasilensis. The other five N2-fixing Paenibacillus strains belonged to cluster III, cluster IV and cluster V (including P. jilunlii, P. zanthoxyli, and P. stellifer mainly).
In this study, 20 of the 25 N2-fixing Paenibacillus strains had inhibitory effects against plant pathogenic fungi, with average inhibition zones varying from 15 to 35 mm on plates. Especially, Paenibacillus sp. SZ-13b could suppress six tested bacterial plant pathogens. Wherease, Paenibacillus sp. SZ-1b, Paenibacillus sp. SZ-15, and Paenibacillus sp. JS-4 could suppress five tested bacterial plant pathogens with strong inhibition activities. The 20 strains with inhibitory effects against plant pathogenic fungi belonged to cluster I and cluster II that were closely related to P. polymyxa, P. jamilae, P. peoriae, and P. brasilensis. Our results are consistent with the previous results that P. polymyxa have long been known for their great ability to produce peptide antibiotics to suppress the growth of plant pathogenic fungi (Deng et al., 2011; He et al., 2007; Helbig, 2001; Raza, Yang & Shen, 2008). For examples, P. polymyxa M1 ( HE577054), which was isolated from root tissues of wheat, was able to promote wheat growth and suppress several phytopathogens (Niu et al., 2011; Yao et al., 2008). P. polymyxa SQR-21 (CP006872) selected from the rhizosphere soil of watermelon could significantly inhibit F. oxysporum (Raza et al., 2009). P. brasilensis PB1 72 (NR025106) isolated from the maize rhizosphere was able to protect seeds and roots against phytopathogenic fungi (Fusarium moniliforme and Diplodia macrospora) (Von der Weid et al., 2005; Von der Weid et al., 2002).
Additionally, 22 N2-fixing Paenibacillus strains (except for Paenibacillus sp. AH-1, Paenibacillus sp. CD-4a and Paenibacillus sp. CD-4b) were capable of producing IAA, which is a primary plant hormone regulating plant growth and development. Among them, Paenibacillus sp. WF-6, Paenibacillus sp. SZ-1a, Paenibacillus sp. SZ-1b, Paenibacillus sp. BJ-5, Paenibacillus sp. YB-3 generated higher yield of IAA.
According to the results of nitrogenase activity, IAA level and inhibitory effect against plant pathogens, 8 strains were chosen to inoculate wheat seedlings, cucumber seedlings and tomato seedlings to analyse their plant promotion effects. We found that Paenibacillus sp. JS-4 and Paenibacillus sp. BJ-4 promoted wheat growth, as well as the chemical nitrogen fertilizer did. While Paenibacillus sp. SZ-10 and Paenibacillus sp. SZ-14 promoted cucumber growth as well as the chemical nitrogen fertilizer did. The two strains Paenibacillus sp. SZ-15 and Paenibacillus sp. BJ-4 significantly promoted tomato growth. Moreover, the 4 strains including Paenibacillus sp. SZ-10, Paenibacillus sp. SZ-14, Paenibacillus sp. YB-10, and Paenibacillus sp. WF-6 could promote tomato growth. From these results, we found that the plant promotion effects exhibited by a Paenibacillus strain varied among plants. At present, we do not know why a same Paenibacillus strain had different promotion effects on different plants.
Taken together, 25 N2-fixning Paenibacillus strains were isolated from plant rhizospheres. The 5 strains including Paenibacillus sp. JS-4, Paenibacillus sp. SZ-10, Paenibacillus sp. SZ-14, Paenibacillus sp. BJ-4 and Paenibacillus sp. SZ-15 with the significant effects of promoting plant growth have great potential as bio-fertilizer.
Microbial fertilizers are widely used in plantation of vegetables in China. The members of Bacillus genus, such as Bacillus subtilis, Bacillus amyloliquefaciens and Bacillus licheniformis, are usually used in biofertilizers. The Paenibacillus strains with nitrogen fixation and multiple bacterial properties for promoting plant growth obtained in this study have great potential to be developed as biofertilizers.
Conclusion
In conclusion, 25 N2-fixing Paenibacillus strains were isolated from plant rhizospheres. Most of them possessed multiple beneficial properties and characteristics of PGPR. They could fix atmospheric nitrogen, produce the profitable phytohormone IAA, control against a wide set of plant pathogens, and enhance growth of diverse important plants. In particular, the five strains including Paenibacillus sp. JS-4, Paenibacillus sp. SZ-10, Paenibacillus sp. SZ-14, Paenibacillus sp. BJ-4 and Paenibacillus sp. SZ-15 with the significant effects of promoting plant growth could be developed and commercially formulated to substitute for environmentally harmful chemical fertilizer and pesticides in field experiments.