A New Geniposidic Acid Derivative Exerts Antiaging Effects through Antioxidative Stress and Autophagy Induction
Abstract
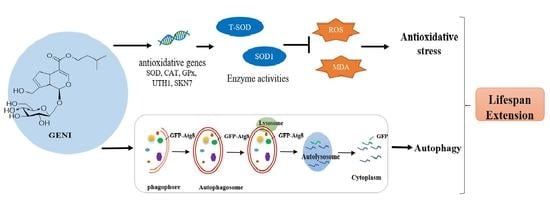
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Isolation of Geniposidic Acid (1) and Geniposide (2)
2.3. Synthesis of Geniposidic Acid Derivatives (3–9)
2.4. Yeast Strains and Lifespan Assay
2.5. Evaluation of Antioxidative Stress
2.6. Measurement of ROS and MDA Levels in Yeast upon Treatment with GENI
2.7. Measurement of SOD, GPx, and CAT Enzymes Activities upon Treatment of GENI in Yeast
2.8. Real-Time Polymerase Chain Reaction (RT-PCR)
2.9. Visualization of Autophagy Induced by GENI in Yeast
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
3.1. Structure Identification of Isolated 1 and 2
3.2. Structure Elucidation of Synthesized GENI
3.3. Structure–Activity Relationship of 1–9
3.4. Extension of the Replicative Lifespan and Chronological Lifespan of K6001 Yeast by GENI
3.5. Increased Activities of SOD, Reduced Contents of ROS and MDA, and Improved Survival Rate of Yeast under Oxidative Stress by GENI
3.6. Effect of GENI on Gene Expression Levels of SOD1, SOD2, GPx and CAT in Yeast
3.7. Involvement of SOD1, SOD2, UTH1, SKN7, CAT, and GPx Genes in the Antiaging Effect of GENI
3.8. Effects of GENI on Autophagy in Yeast
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| GENI | Geniposidic 4-isoamyl ester |
| CAT | Catalase |
| SOD | Superoxide dismutase |
| ROS | Reactive oxygen species |
| MDA | Malondialdehyde |
| GPx | Glutathione peroxidase |
| GFP | Green fluorescent protein |
| RES | Resveratrol |
References
- United Nations. Department of Economic and Social Affairs, Population Division. World Population Ageing 2019. Available online: https://www.un.org/development/desa/pd/zh/node/3172 (accessed on 31 December 2019).
- Mitchell, S.J.; Knudsen, M.S.; Longo, D.L.; Cabo, R. Animal models of aging research: Implications for human aging and age-related diseases. Annu. Rev. Anim. Biosci. 2015, 3, 283–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [Green Version]
- Drose, S.; Brandt, U. Molecular Mechanisms of Superoxide Production by the Mitochondrial Respiratory Chain. In Mitochondrial Oxidative Phosphorylation; Kadenbach, B., Ed.; Springer Science Business Media: New York, NY, USA, 2012; pp. 145–170. [Google Scholar]
- Mockett, R.J.; Sohal, R.S.; Orr, W.C. Overexpression of glutathione reductase extends survival in transgenic Drosophila melanogaster under hyperoxia but not normoxia. FASEB J. 1999, 13, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Orr, W.C.; Sohal, R.S. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science 1994, 263, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Autophagy revisited: A conversation with christian de duve. Autophagy 2008, 4, 740–743. [Google Scholar] [CrossRef] [Green Version]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian autophagy: How does it work. Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef]
- Osawa, T.; Ishii, Y.; Noda, N.N. Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells 2020, 25, 65–70. [Google Scholar] [CrossRef]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, A.F.; Sebti, S.; Wei, Y.J.; Zou, Z.J.; Shi, M.J.; McMillan, K.L.; He, C.C.; Ting, T.; Liu, Y.; Chiang, W.-C.; et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef]
- Barton, A.A. Some aspects of cell division in Saccharomyces cerevisiae. J. Gen. Microbiol. 1950, 4, 84–86. [Google Scholar] [CrossRef] [Green Version]
- Marques, B.S.; Burhans, W.C.; Ludovico, P. Yeast at the Forefront of Research on Ageing and Age-Related Diseases. In Yeasts in Biotechnology and Human Health; Correia, I.S., Ed.; Springer Nature Switzerland AG: Gewerbestrasse, Switzerland, 2019; pp. 217–242. [Google Scholar]
- Kaeberlein, M. Resveratrol and rapamycin: Are they anti-aging drugs. BioEssays 2010, 32, 96–99. [Google Scholar] [CrossRef]
- Chen, L.P.; Li, M.X.; Yang, Z.Q.; Tao, W.D.; Wang, P.; Tian, X.Y.; Li, X.L.; Wang, W.G. Gardenia jasminoides Ellis: Ethnopharmacology, phytochemistry, and pharmacological and industrial applications of an important traditional Chinese medicine. J. Ethnopharmacol. 2020, 257, 112829. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Zhang, R.Q.; Rahman, K.; Cao, Z.X.; Zhang, H.; Peng, C. Diverse pharmacological activities and potential medicinal benefits of geniposide. Evid-Based Complement. Altern. Med. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.H.; Zhang, H.J.; Li, H.; Lv, C.; Liu, X.L.; Li, Z.; Xin, W.F.; Wang, Y.Y.; Zhang, W.S. Geniposide ameliorates cognitive deficits by attenuating the cholinergic defect and amyloidosis in middle-aged Alzheimer model mice. Neuropharmacology 2017, 116, 18–29. [Google Scholar] [CrossRef]
- Güvenalp, Z.; Kilic, N.; Kazaz, C.; Kaya, Y.; Demirezer, L.O. Chemical constituents of Galium tortumense. Turk. J. Chem. 2006, 30, 515–523. [Google Scholar]
- Lin, Y.F.; Kotakeyama, Y.; Li, J.; Pan, Y.J.; Matsuura, A.; Ohya, Y.; Yoshida, M.; Xiang, L.; Qi, J.H. Cucurbitacin B exerts antiaging effects in yeast by regulating autophagy and oxidative stress. Oxidative Med. Cell. Longev. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Liu, Y.N.; Fujii, R.; Farooq, U.; Cheng, L.H.; Matsuura, A.; Qi, J.H.; Xiang, L. Ehretiquinone from onosma bracteatum wall exhibits antiaging effect on yeasts and mammals through antioxidative stress and autophagy induction. Oxidative Med. Cell. Longev. 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Disasa, D.; Cheng, L.H.; Manzoor, M.; Liu, Q.; Wang, Y.; Xiang, L.; Qi, J.H. Amarogentin from Gentiana rigescens Franch exhibits antiaging and neuroprotective effects through antioxidative stress. Oxidative Med. Cell. Longev. 2020, 2020, 1–15. [Google Scholar] [CrossRef]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Jiang, T.F.; Sun, Q.; Chen, S.D. Oxidative stress: A major pathogenesis and potential therapeutic target of antioxidative agents in Parkinson’s disease and Alzheimer’s disease. Prog. Neurobiol. 2016, 147, 1–19. [Google Scholar] [CrossRef]
- Kirkin, V. History of the selective autophagy research: How did it begin and where does it stand today. J. Mol. Biol. 2020, 432, 3–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cheng, L.H.; Matsuura, A.; Xiang, L.; Qi, J.H. Gentiopicroside, a secoiridoid glycoside from Gentiana rigescens Franch, extends the lifespan of yeast via inducing mitophagy and antioxidative Stress. Oxidative Med. Cell. Longev. 2020, 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Pan, Y.; Liu, Y.; Disasa, D.; Akira, M.; Xiang, L.; Qi, J. A New Geniposidic Acid Derivative Exerts Antiaging Effects through Antioxidative Stress and Autophagy Induction. Antioxidants 2021, 10, 987. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060987
Wang Y, Pan Y, Liu Y, Disasa D, Akira M, Xiang L, Qi J. A New Geniposidic Acid Derivative Exerts Antiaging Effects through Antioxidative Stress and Autophagy Induction. Antioxidants. 2021; 10(6):987. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060987
Chicago/Turabian StyleWang, Ying, Yanjun Pan, Yanan Liu, Dejene Disasa, Matsuura Akira, Lan Xiang, and Jianhua Qi. 2021. "A New Geniposidic Acid Derivative Exerts Antiaging Effects through Antioxidative Stress and Autophagy Induction" Antioxidants 10, no. 6: 987. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10060987