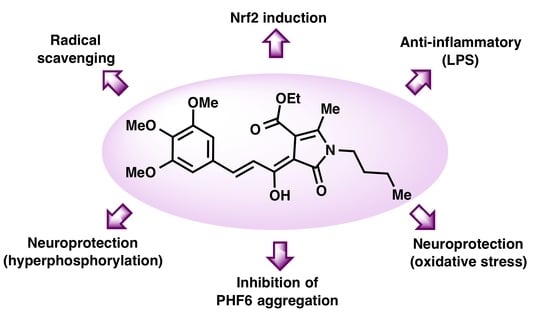
Curcumin-Piperlongumine Hybrids with a Multitarget Profile Elicit Neuroprotection in In Vitro Models of Oxidative Stress and Hyperphosphorylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. General Experimental Details
2.1.2. General Procedure for the Synthesis of 2-Pyrrolin-5-one derivatives 1
2.1.3. Synthesis of Compounds 2
2.1.4. Prediction of Physicochemical, ADME and CNS Permeability Properties
2.1.5. Antioxidant Capacity by the ORAC Assay
2.1.6. Anti-Oxidant Capacity by the 1,1-Diphenyl-2-picryl-hydrazyl (DPPH) Reduction Assay
2.1.7. Computational Study of PHF6 Hexapeptide Aggregation
2.2. Biological Studies
2.2.1. Culture of HEK293-Tau3R Cells
2.2.2. Culture of SH-SY5Y Neuroblastoma Cells
2.2.3. Culture of AREc32 Cells
2.2.4. Culture of BV2 Cells
2.2.5. Cell Viability Studies in HEK293-Tau3R and SH-SY5Y Cells
2.2.6. Reactive Oxygen Species (ROS) Measurement in HEK293-Tau3R Cells
2.2.7. PHF6 Peptide Aggregation
2.2.8. Thioflavin T Assay
2.2.9. Nrf2 Induction Capacity
2.2.10. Neuroprotection Assays in SH-SY5Y Cells
2.2.11. Nitrite Production Reduction Assay
2.3. Statistical Analyses
3. Results and Discussion
3.1. Prediction of Physicochemical, ADME and CNS Permeability Properties of Compounds 2
3.2. Synthesis of Compounds 2
3.3. In Vitro Characterization of the Antioxidant Capacity of Compounds 2
3.4. Biological Evaluation of Curcumin Derivatives
3.4.1. Cytotoxicity Evaluation in the HEK293-Tau3R and SH-SY5Y Cell Lines
3.4.2. ROS Scavenger Activity in HEK293-Tau3R Cells
3.4.3. Nrf2 Induction
3.4.4. Inhibition of PHF6 Aggregation
3.4.5. Anti-Inflammatory Properties
3.4.6. Neuroprotection against Oxidative Stress
3.4.7. Neuroprotection against Tau Hyperphosphorylation Induced by Okadaic Acid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Guo, C.; Kong, J. Oxidative stress in neurodegenerative diseases. Neural Regen. Res. 2012, 7, 376–385. [Google Scholar]
- Muralidar, S.; Ambi, S.V.; Sekaran, S.; Thirumalai, D.; Palaniappan, B. Role of tau protein in Alzheimer’s disease: The prime pathological player. Int. J. Biol. Macromol. 2020, 163, 1599–1617. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Li, P.; Wei, N.; Zhao, Z.; Liang, H.; Ji, X.; Chen, W.; Xue, M.; Wei, J. The ambiguous relationship of oxidative stress, tau hyperphosphorylation, and autophagy dysfunction in Alzheimer’s disease. Oxid. Med. Cell. Longev. 2015, 2015, 352723. [Google Scholar] [CrossRef]
- Haque, M.M.; Murale, D.J.; Kim, Y.K.; Lee, J.S. Crosstalk between oxidative stress and tauopathy. Int. J. Mol. Sci. 2019, 20, 1959. [Google Scholar] [CrossRef] [Green Version]
- Nizynski, B.; Dzwolak, W.; Nieznanski, K. Amyloidogenesis of Tau protein. Protein Sci. 2017, 26, 2126–2150. [Google Scholar] [CrossRef] [Green Version]
- Sinsky, J.; Pichlerova, K.; Hanes, J. Tau protein interaction partners and their roles in Alzheimer’s disease and other tauopathies. Int. J. Mol. Sci. 2021, 22, 9207. [Google Scholar] [CrossRef]
- Rojas-Quijano, F.A.; Morrow, D.; Wise, B.M.; Brancia, F.L.; Goux, W.J. Prediction of nucleating sequences from amyloidogenic propensities of tau-related peptides. Biochemistry 2006, 45, 4638–4652. [Google Scholar] [CrossRef]
- Cores, Á.; Piquero, M.; Villacampa, M.; León, R.; Menéndez, J.C. NRF2 regulation processes as a source of potential drug targets against neurodegenerative diseases. Biomolecules 2020, 10, 904. [Google Scholar] [CrossRef]
- Prasad, S.; Aggarwal, B.B. Turmeric, the golden spice. In Herbal Medicine: Biomolecular and Clinical Aspects, 2nd ed.; Benzie, I.F.F., Wachtel-Galor, S., Eds.; CRS Press/Taylor and Francis: Boca Raton, FL, USA, 2011. [Google Scholar]
- Peng, Y.; Ao, M.; Dong, B.; Jiang, Y.; Yu, L.; Chen, Z.; Hu, C.; Xu, R. Anti-Inflammatory effects of curcumin in inflammatory diseases: Status, limitations and countermeasures. Drug Des. Devel. Ther. 2021, 15, 4503–4525. [Google Scholar] [CrossRef]
- Tizabi, Y.; Hurley, L.L.; Qualls, Z.; Akinfiresoye, L. Relevance of the Anti-Inflammatory Properties of Curcumin in Neurodegenerative Diseases and Depression. Molecules 2014, 19, 20864–20879. [Google Scholar] [CrossRef]
- Lee, W.H.; Loo, C.Y.; Bebawy, M.; Luk, F.; Mason, R.S.; Rohanizadeh, R. Curcumin and its derivatives: Their application in neuropharmacology and neuroscience in the 21st century. Curr. Neuropharmacol. 2013, 11, 338–378. [Google Scholar] [CrossRef] [Green Version]
- Farooqui, A.A. Therapeutic Potentials of Curcumin for Alzheimer Disease; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Shin, J.W.; Chun, K.; Kim, D.H.; Kim, S.J.; Kim, S.H.; Cho, N.C.; Na, H.K.; Surh, Y.J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef]
- Soeda, Y.; Takashima, A. New insights into drug discovery targeting Tau protein. Front. Mol. Neurosci. 2020, 13, 590896. [Google Scholar] [CrossRef]
- Rane, J.S.; Bhaumik, P.; Panda, D. Curcumin inhibits tau aggregation and disintegrates preformed tau filaments in vitro. J. Alzheimer’s Dis. 2017, 60, 999–1014. [Google Scholar] [CrossRef]
- Bijari, N.; Balalaie, S.; Akbari, V.; Golmohammadi, F.; Moradi, S.; Adibi, H.; Khodarahmi, R. Effective suppression of the modified PHF6 peptide/1N4R Tau amyloid aggregation by intact curcumin, not its degradation products: Another evidence for the pigment as preventive/therapeutic “functional food”. Int. J. Biol. Macromol. 2018, 120, 1009–1022. [Google Scholar] [CrossRef]
- Askarizadeha, A.; Barreto, G.E.; Henney, N.C.; Majeed, M.; Sahebkar, A. Neuroprotection by curcumin: A review on brain delivery strategies. Int. J. Pharm. 2020, 585, 119476. [Google Scholar] [CrossRef]
- Vareed, S.K.; Kakarala, M.; Ruffin, M.T.; Crowell, J.A.; Normolle, D.P.; Djuric, Z.; Brenner, D.E. Pharmacokinetics of curcumin conjugate metabolites in healthy human subjects. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1411–1417. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.; Do, J.; Jae-sung, B.; Jin, H.K.; Kim, J.-H.; Inn, K.-S.; Oh, M.S.; Lee, J.K. Piperlongumine inhibits neuroinflammation via regulating NF-κB signaling pathways in lipopolysaccharide-stimulated BV2 microglia cells. J. Pharmacol. Sci. 2018, 137, 195–201. [Google Scholar] [CrossRef]
- Li, G.; Zheng, Y.; Yao, J.; Hu, L.; Liu, Q.; Ke, F.; Feng, W.; Zhao, Y.; Yan, P.; He, W.; et al. Design and green synthesis of piperlongumine analogs and their antioxidant activity against cerebral ischemia-reperfusion injury. ACS Chem. Neurosci. 2019, 10, 4545–4557. [Google Scholar] [CrossRef]
- Rampa, A.; Montanari, S.; Pruccoli, L.; Bartolini, M.; Falchi, F.; Feoli, A.; Cavalli, A.; Belluti, F.; Gobbi, S.; Tarozzi, A.; et al. Chalcone-based carbamates for Alzheimer’s disease treatment. Future Med. Chem. 2017, 9, 749–764. [Google Scholar] [CrossRef]
- Cores, A.; Abril, S.; Michalska, P.; Duarte, P.; Olives, A.I.; Martín, M.A.; Villacampa, M.; León, R.; Menéndez, J.C. Bisavenathramide analogues as Nrf2 inductors and neuroprotectors in in vitro models of oxidative stress and hyperphosphorylation. Antioxidants 2021, 10, 941. [Google Scholar] [CrossRef]
- Cores, A.; Estévez, V.; Villacampa, M.; Menéndez, J.C. Three-component access to 2-pyrrolin-5-ones and their use in target-oriented and diversity-oriented synthesis. RSC Adv. 2016, 6, 39433–39443. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Release 2021-3: QikProp; Schrödinger, LLC: New York, NY, USA, 2021.
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-3: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021.
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Moving beyond rules: The development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 2010, 1, 435–449. [Google Scholar] [CrossRef] [Green Version]
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Central nervous system multiparameter optimization desirability: Application in drug discovery. ACS Chem. Neurosci. 2016, 7, 767–775. [Google Scholar] [CrossRef] [Green Version]
- Rankovic, Z. CNS physicochemical property space shaped by a diverse set of molecules with experimentally determined exposure in the mouse brain. J. Med. Chem. 2017, 60, 5943–5954. [Google Scholar] [CrossRef]
- Herrera-Arozamena, C.; Estrada-Valencia, M.; Pérez, C.; Lagartera, L.; Morales-García, J.A.; Pérez-Castillo, A.; Franco-González, J.F.; Michalska, P.; Duarte, P.; León, R.; et al. Tuning melatonin receptor subtype selectivity in oxadiazolone-based analogues: Discovery of QR2 ligands and NRF2 activators with neurogenic properties. Eur. J. Med. Chem. 2020, 190, 112090. [Google Scholar] [CrossRef]
- Davalos, A.; Gómez-Cordoves, C.; Bartolomé, B. Extending applicability of the oxygen radical absorbance capacity (ORAC-fluorescein) assay. J. Agric. Food Chem. 2004, 52, 48–54. [Google Scholar] [CrossRef]
- Dudonne, S.; Vitrac, X.; Coutiere, P.; Woillez, M.; Merillon, J.M. Comparative study of antioxidant properties and total phenolic content of 30 plant extracts of industrial interest using DPPH, ABTS, FRAP, SOD, and ORAC assays. J. Agric. Food Chem. 2009, 57, 1768–1774. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2012, 31, 455–461. [Google Scholar]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B.; GROMACS Development Team. GROMACS User Manual Version 2018.4. Available online: www.gromacs.org (accessed on 16 May 2021).
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force field. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; He, X.; Vanommeslaeghe, K.; MacKerell, A.D., Jr. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012, 33, 2451–2468. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM general force field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://cgenff.umaryland.edu (accessed on 15 July 2021).
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- LeBel, C.P.; Ischiropoulos, H.; Bondy, S.C. Evaluation of the probe 2′,7′-dichlorofluorescein as an indicator of reactive oxygen species formation and oxidative stress. Chem. Res. Toxicol. 1992, 5, 227–231. [Google Scholar] [CrossRef] [Green Version]
- Lopresti, A.L. The problem of curcumin and its bioavailability: Could its gastrointestinal influence contribute to its overall health-enhancing effects? Adv. Nutr. 2018, 9, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.-M.; Chien, C.F.; Lin, L.-C.; Tsai, T.-H. Curcumin and its nano-formulation: The kinetics of tissue distribution & blood-brain-barrier penetration. Int. J. Pharm. 2011, 416, 331–338. [Google Scholar] [PubMed]
- Lee, E.H.-C.; Lim, S.S.-C.; Yuen, K.-H.; Lee, C.-Y. Curcumin and a hemi-analogue with improved blood–brain barrier permeability protect against amyloid-beta toxicity in Caenorhabditis elegans via SKN-1/Nrf activation. J. Pharm. Pharmacol. 2019, 71, 860–868. [Google Scholar] [CrossRef] [PubMed]
- García-Alloza, M.; Borrelli, L.A.; Rozkalne, A.; Hyman, B.T.; Bacskai, B.J. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J. Neurochem. 2007, 102, 1095–1104. [Google Scholar] [CrossRef]
- Mohammadkhani, L.; Heravi, M.M. Oxalyl chloride: A versatile reagent in organic transformations. ChemistrySelect 2019, 4, 6309–6337. [Google Scholar] [CrossRef]
- Borra, S.K.; Gurumurthy, P.; Mahendra, J.; Ch, R. Antioxidant and free radical scavenging activity of curcumin determined by using different in vitro and ex vivo models. J. Med. Plant. Res. 2013, 7, 2680–2690. [Google Scholar]
- Choudhury, A.K.; Raja, S.; Mahapatra, S.; Nagabhushanam, K.; Majeed, M. Synthesis and evaluation of the anti-oxidant capacity of curcumin glucuronides, the major curcumin metabolites. Antioxidants 2015, 4, 750–767. [Google Scholar] [CrossRef] [Green Version]
- Apak, R.; Ozyurek, M.; Guclu, K.; Capanoglu, E. Antioxidant activity/capacity measurement. 2. Hydrogen Atom Transfer (HAT)-based, mixed-mode (electron transfer (ET)/HAT), and lipid peroxidation assays. J. Agric. Food Chem. 2016, 64, 1028–1045. [Google Scholar] [CrossRef]
- Houck, A.L.; Hernández, F.; Ávila, J. A simple model to study tau pathology. J. Exp. Neurosci. 2016, 10, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Ashrafizadeh, M.; Ahmadi, Z.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Curcumin activates the Nrf2 pathway and induces cellular protection against oxidative injury. Curr. Mol. Med. 2020, 20, 116–133. [Google Scholar] [CrossRef]
- Sarkar, B.; Dhiman, M.; Mittal, S.; Mantha, A.K. Curcumin revitalizes Amyloid beta (25-35)-induced and organophosphate pesticides pestered neurotoxicity in SH-SY5Y and IMR-32 cells via activation of APE1 and Nrf2. Metab. Brain Dis. 2017, 32, 2045–2061. [Google Scholar] [CrossRef]
- Wang, X.J.; Hayes, J.D.; Wolf, C.R. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of Nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006, 66, 10983–10994. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.C.; McDonald, P.R.; Liu, J.; Klaassen, C.D. Screening of natural compounds as activators of the Keap1-Nrf2 pathway. Planta Med. 2014, 80, 97–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, A.; Viswanathan, G.K.; Huber, A.; Arad, E.; Engel, H.; Jelinek, R.; Gazit, E.; Segal, D. Inhibition of tau amyloid formation and disruption of its preformed fibrils by naphthoquinone-dopamine hybrid. FEBS J. 2021, 288, 4267–4290. [Google Scholar] [CrossRef]
- Rawat, N.; Biswas, P. Hydrogen bond dynamics in intrinsically disordered proteins. J. Phys. Chem. B 2014, 118, 3018–3025. [Google Scholar] [CrossRef]
- Smit, F.X.; Luiken, J.A.; Bolhuis, P.G. Primary fibril nucleation of aggregation prone tau fragments PHF6 and PHF6*. J. Phys. Chem. B 2017, 121, 3250–3261. [Google Scholar] [CrossRef]
- Landau, M.; Sawaya, M.R.; Faull, K.F.; Laganowsky, A.; Jiang, L.; Sievers, S.A.; Liu, J.; Barrio, J.R.; Eisenberg, D. Towards a pharmacophore for amyloid. PLoS Biol. 2011, 9, 25–27. [Google Scholar] [CrossRef]
- Gazit, E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002, 16, 77–83. [Google Scholar] [CrossRef]
- Brahmachari, S.; Paul, A.; Segal, D.; Gazit, E. Inhibition of amyloid oligomerization into different supramolecular architectures by small molecules: Mechanistic insights and design rules. Future Med. Chem. 2017, 9, 797–810. [Google Scholar] [CrossRef]
- Krishna Kumar, V.G.; Paul, A.; Gazit, E.; Segal, D. Mechanistic insights into remodeled Tau-derived PHF6 peptide fibrils by naphthoquinone-tryptophan hybrids. Sci. Rep. 2018, 8, 71. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Gyengesi, E.; Munch, G. In search of an anti-inflammatory drug for Alzheimer disease. Nat. Rev. Neurol. 2020, 16, 131–132. [Google Scholar] [CrossRef]
- Heneka, M.T.; Golenbock, D.T.; Latz, E. Innate immunity in Alzheimer’s disease. Nat. Immunol. 2015, 16, 229–236. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, J.B.; Moore, K.J.; Means, T.K.; Leung, J.; Terada, K.; Toft, M.; Freeman, M.W.; Luster, A.D. CD36 mediates the innate host response to beta-amyloid. J. Exp. Med. 2003, 197, 1657–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneka, M.T.; Kummer, M.P.; Stutz, A.; Delekate, A.; Schwartz, S.; Vieira-Saecker, A.; Griep, A.; Axt, D.; Remus, A.; Tzeng, T.C.; et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 2013, 493, 674–678. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2, e17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, W.; Sun, C.; Ma, Y.; Wang, S.; Wang, X.; Zhang, Y. Inhibition of TLR4 induces M2 microglial polarization and provides neuroprotection via the NLRP3 inflammasome in Alzheimer’s disease. Front. Neurosci. 2020, 14, 444. [Google Scholar] [CrossRef]
- Gehrmann, J.; Matsumoto, Y.; Kreutzberg, G.W. Microglia: Intrinsic immuneffector cell of the brain. Brain Res. Rev. 1995, 20, 269–287. [Google Scholar] [CrossRef]
- Subhramanyam, C.S.; Wang, C.; Hu, Q.; Dheen, S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin. Cell Dev. Biol. 2019, 94, 112–120. [Google Scholar] [CrossRef]
- Yuste, J.E.; Tarragón, E.; Campuzano, C.M.; Ros-Bernal, F. Implications of glial nitric oxide in neurodegenerative diseases. Front. Cell. Neurosci. 2015, 9, 322. [Google Scholar] [CrossRef] [Green Version]
- Michalska, P.; Buendia, I.; Duarte, P.; Fernández-Mendívil, C.; Negredo, P.; Cuadrado, A.; López, M.G.; León, R. Melatonin-sulforaphane hybrid ITH12674 attenuates glial response in vivo by blocking LPS binding to MD2 and receptor oligomerization. Pharmacol. Res. 2020, 152, 104597. [Google Scholar] [CrossRef]
- Sharma, C.; Kim, S.R. Linking oxidative stress and proteinopathy in Alzheimer’s disease. Antioxidants 2021, 10, 1231. [Google Scholar] [CrossRef] [PubMed]
- Michalska, P.; León, R. When it comes to an end: Oxidative stress crosstalk with protein aggregation and neuroinflammation induce neurodegeneration. Antioxidants 2020, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Tenti, G.; Parada, E.; León, R.; Egea, J.; Martínez-Revelles, S.; Briones, A.M.; Sridharan, V.; López, M.G.; Ramos, M.T.; Menéndez, J.C. New 5-unsubstituted dihydropyridines with improved CaV1.3 selectivity as potential neuroprotective agents against ischemic injury. J. Med. Chem. 2014, 57, 4313–4323. [Google Scholar] [CrossRef] [PubMed]
- Michalska, P.; Mayo, P.; Fernández-Mendívil, C.; Tenti, G.; Duarte, P.; Buendia, I.; Ramos, M.T.; López, M.G.; Menéndez, J.C.; León, R. Antioxidant, anti-inflammatory and neuroprotective profiles of novel 1,4-dihydropyridine derivatives for the treatment of Alzheimer’s disease. Antioxidants 2020, 9, 650. [Google Scholar] [CrossRef]
- Michalska, P.; Tenti, G.; Satriani, M.; Cores, A.; Ramos, M.T.; García, A.G.; Menéndez, J.C.; León, R. Aza-CGP37157-lipoic hybrids designed as novel Nrf2-inducers and antioxidants exert neuroprotection against oxidative stress and show neuroinflammation inhibitory properties. Drug. Dev. Res. 2020, 81, 283–294. [Google Scholar] [CrossRef]
- Qualls, Z.; Brown, D.; Ramlochansingh, C.; Hurley, L.L.; Tizabi, Y. Protective effects of curcumin against rotenone and salsolinol-induced toxicity: Implications for Parkinson’s disease. Neurotox Res. 2014, 25, 81–89. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Yang, D.; Smith, W.W. Curcumin protects against rotenone-induced neurotoxicity in cell and Drosophila models of Parkinson’s disease. Adv. Parkinsons Dis. 2013, 2, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Buratta, S.; Chiaradia, E.; Tognoloni, A.; Gambelunghe, A.; Meschini, C.; Palmieri, L.; Muzi, G.; Urbanelli, L.; Emiliani, C.; Tancini, B. Effect of curcumin on protein damage induced by rotenone in dopaminergic PC12 cells. Int. J. Mol. Sci. 2020, 21, 2761. [Google Scholar] [CrossRef] [PubMed]
- Parada, E.; Buendia, I.; Navarro, E.; Avendaño, C.; Egea, J.; López, M.G. Microglial HO-1 induction by curcumin provides antioxidant, antineuroinflammatory, and glioprotective effects. Mol. Nutr. Food Res. 2015, 59, 1690–1700. [Google Scholar] [CrossRef]
- Lovell, M.A.; Xiong, S.; Xie, C.; Davies, P.; Markesbery, W.R. Induction of hyperphosphorylated tau in primary rat cortical neuron cultures mediated by oxidative stress and glycogen synthase kinase-3. J. Alzheimers Dis. 2004, 6, 659–671. [Google Scholar] [CrossRef]
- Alavi Naini, S.M.; Soussi-Yanicostas, N. Tau hyperphosphorylation and oxidative stress, a critical vicious circle in neurodegenerative tauopathies? Oxid. Med. Cell Longev. 2015, 2015, 151979. [Google Scholar] [CrossRef] [Green Version]
- Luengo, E.; Buendia, I.; Fernández-Mendívil, C.; Trigo-Alonso, P.; Negredo, P.; Michalska, P.; Hernández-García, B.; Sánchez-Ramos, C.; Bernal, J.A.; Ikezu, T.; et al. Pharmacological doses of melatonin impede cognitive decline in tau-related Alzheimer models, once tauopathy is initiated, by restoring the autophagic flux. J. Pineal Res. 2019, 67, e12578. [Google Scholar] [CrossRef]
- Gameiro, I.; Michalska, P.; Tenti, G.; Cores, A.; Buendia, I.; Rojo, A.I.; Georgakopoulos, N.D.; Hernández-Guijo, J.M.; Ramos, M.T.; Wells, G.; et al. Discovery of the first dual GSK3beta inhibitor/Nrf2 inducer. A new multitarget therapeutic strategy for Alzheimer’s disease. Sci. Rep. 2017, 7, 45701. [Google Scholar] [CrossRef]
- Ravindran, J.; Gupta, N.; Agrawal, M.; Bala Bhaskar, A.S.; Lakshmana Rao, P.V. Modulation of ROS/MAPK signaling pathways by okadaic acid leads to cell death, via mitochondrial mediated caspase-dependent mechanism. Apoptosis 2011, 16, 145–161. [Google Scholar] [CrossRef]
- Rajasekar, N.; Dwivedi, S.; Kumar, S.; Pradeep, T.; Kamat, K.; Hanif, K.; Nath, C.; Shukla, R. Neuroprotective effect of curcumin on okadaic acid induced memory impairment in mice. Eur. J. Pharmacol. 2013, 715, 381–394. [Google Scholar] [CrossRef]










| Cmpd. | MW (g/mol) | TPSA (Å2) | Heavy Atoms | HBA | HBD | RotB | Log P | QPP Caco (nm/s) | QPP MDCK (nm/s) | %Human Oral Absorption | Rule of Five |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Curcumin | 368.38 | 96.22 | 27 | 6 | 3 | 12 | 3.17 | 141.2 | 59.6 | 81.8 | 0 |
| 2a | 497.51 | 94.53 | 36 | 8 | 1 | 12 | 4.09 | 903.3 | 799.9 | 100 | 1 |
| 2b | 431.48 | 94.53 | 31 | 7 | 1 | 12 | 3.13 | 864.1 | 422.5 | 96.5 | 1 |
| 2c | 483.49 | 94.53 | 35 | 8 | 1 | 11 | 3.71 | 428.8 | 357.5 | 95.8 | 1 |
| 2d | 445.51 | 94.53 | 32 | 7 | 1 | 13 | 3.47 | 1044.3 | 518.4 | 100 | 1 |
| 2e | 387.43 | 96.3 | 28 | 6 | 2 | 11 | 2.69 | 314.4 | 141.6 | 100 | 0 |
| 2f | 465.50 | 94.53 | 34 | 7 | 1 | 11 | 3.36 | 980.3 | 484.2 | 100 | 1 |
| Entry | Compound | CNS MPO.v2 [0–6] |
|---|---|---|
| 1 | Curcumin | 3.97 |
| 2 | 2a | 4.03 |
| 3 | 2b | 4.93 |
| 4 | 2c | 4.23 |
| 5 | 2d | 4.63 |
| 6 | 2e | 4.60 |
| 7 | 2f | 4.53 |
| Entry | Compound | ORAC | DPPH |
|---|---|---|---|
| Trolox Eqs. | SC50, μM | ||
| 1 | Curcumin | 3.88 ± 0.18 | 18.2 ± 0.63 |
| 2 | Ascorbic acid | -- | 17.2 ± 1.14 |
| 3 | 2a | 0.50 ± 0.23 *** | 20.6 ± 2.17 |
| 4 | 2b | 0.52 ± 0.22 *** | 10.8 ± 0.06 ***/### |
| 5 | 2c | 0.35 ± 0.13 *** | 56.9 ± 1.18 **/## |
| 6 | 2d | 1.63 ± 0.06 *** | 14.2 ± 2.15 |
| 7 | 2e | 1.73 ± 0.68 *** | 8.87 ± 0.45 ***/### |
| 8 | 2f | 0.65 ± 0.11 *** | 36.6 ± 1.02 |
| Parameter | Apo-System | With Curcumin | With Compound 2d |
|---|---|---|---|
| RMSD (Å) | 1.16 ± 0.33 | 1.85 ± 0.35 | 2.47 ± 0.80 |
| Rg | 1.94 ± 0.19 | 2.33 ± 0.24 | 3.00 ± 0.60 |
| Hbond | 122.18 ± 6.13 | 118.23 ± 5.55 | 112.02 ± 4.70 |
| Entry | Compound | IC50 (μM) BV2 |
|---|---|---|
| 1 | Curcumin | 0.64 ± 0.17 |
| 2 | 2a | 0.30 ± 0.04 * |
| 3 | 2b | 0.42 ± 0.08 |
| 4 | 2c | 0.56 ± 0.03 |
| 5 | 2d | 0.55 ± 0.08 |
| 6 | 2e | 0.37 ± 0.05 |
| 7 | 2f | 0.76 ± 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cores, Á.; Carmona-Zafra, N.; Martín-Cámara, O.; Sánchez, J.D.; Duarte, P.; Villacampa, M.; Bermejo-Bescós, P.; Martín-Aragón, S.; León, R.; Menéndez, J.C. Curcumin-Piperlongumine Hybrids with a Multitarget Profile Elicit Neuroprotection in In Vitro Models of Oxidative Stress and Hyperphosphorylation. Antioxidants 2022, 11, 28. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11010028
Cores Á, Carmona-Zafra N, Martín-Cámara O, Sánchez JD, Duarte P, Villacampa M, Bermejo-Bescós P, Martín-Aragón S, León R, Menéndez JC. Curcumin-Piperlongumine Hybrids with a Multitarget Profile Elicit Neuroprotection in In Vitro Models of Oxidative Stress and Hyperphosphorylation. Antioxidants. 2022; 11(1):28. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11010028
Chicago/Turabian StyleCores, Ángel, Noelia Carmona-Zafra, Olmo Martín-Cámara, Juan Domingo Sánchez, Pablo Duarte, Mercedes Villacampa, Paloma Bermejo-Bescós, Sagrario Martín-Aragón, Rafael León, and J. Carlos Menéndez. 2022. "Curcumin-Piperlongumine Hybrids with a Multitarget Profile Elicit Neuroprotection in In Vitro Models of Oxidative Stress and Hyperphosphorylation" Antioxidants 11, no. 1: 28. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11010028