Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2
Abstract
:1. Introduction
2. The Role of Nrf2 to Maintain Redox Homeostasis in Cardiometabolic Diseases
Nrf2 Structure and Regulatory Mechanisms
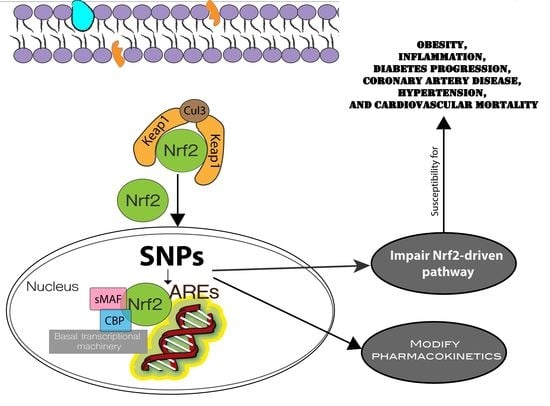
3. Nrf2 Genetic Variations (Single Nucleotide Polymorphisms)
4. Contribution of Nrf2/Keap1 and Target Genes Polymorphisms in Cardiometabolic Diseases
4.1. Obesity and Diabetes
4.2. Coronary Artery Disease (CAD)
4.3. Hypertension
5. Implications of Nrf2-Related Genes Polymorphisms in Cardiovascular Drug Therapy
6. Coffee Consumption, Genetic Polymorphisms of Nrf2 and Cardiometabolic Diseases
7. Association of Environmental Factors with Nrf2 and Related Genes Expression in Cardiometabolic Diseases
8. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Cardiometabolic Diseases. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 28 January 2022).
- Barteková, M.; Adameová, A.; Görbe, A.; Ferenczyová, K.; Pecháňová, O.; Lazou, A.; Dhalla, N.S.; Ferdinandy, P.; Giricz, Z. Natural and synthetic antioxidants targeting cardiac oxidative stress and redox signaling in cardiometabolic diseases. Free Radic. Biol. Med. 2021, 169, 446–477. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Mann, G.E.; Chapple, S.J. Concerted redox modulation by sulforaphane alleviates diabetes and cardiometabolic syndrome. Free Radic. Biol. Med. 2018, 122, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Yoh, K.; Kobayashi, A.; Ishii, Y.; Kure, S.; Koyama, A.; Sakamoto, T.; Sekizawa, K.; Motohashi, H.; Yamamoto, M. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem. Biophys. Res. Commun. 2004, 321, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.-C.; Chang, L.-C.; Tseng, J.-C.; Chu, C.-M.; Liu, Y.-C.; Shieh, W.-B. Functional haplotypes in the promoter region of transcription factor Nrf2 in chronic obstructive pulmonary disease. Dis. Markers 2010, 28, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-Y.; Marzec, J.; Kleeberger, S.R. Functional polymorphisms in Nrf2: Implications for human disease. Free Radic. Biol. Med. 2015, 88, 362–372. [Google Scholar] [CrossRef]
- Villalobos, J.B.; Molina-Muñoz, T.; Mailloux-Salinas, P.; Bravo, G.; Carvajal, K.; Gómez-Víquez, N.L. Oxidative stress in cardiomyocytes contributes to decreased SERCA2a activity in rats with metabolic syndrome. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H1344–H1353. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Camacho, I.; García-Niño, W.; Flores-García, M.; Pedraza-Chaverri, J.; Zazueta, C. Alteration of mitochondrial supercomplexes assembly in metabolic diseases. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165935. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef]
- Lee, Y.; Im, E. Regulation of miRNAs by Natural Antioxidants in Cardiovascular Diseases: Focus on SIRT1 and eNOS. Antioxidants 2021, 10, 377. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef]
- Ibrahim, L.; Mesgarzadeh, J.; Xu, I.; Powers, E.T.; Wiseman, R.L.; Bollong, M.J. Defining the Functional Targets of Cap‘n’collar Transcription Factors NRF1, NRF2, and NRF3. Antioxidants 2020, 9, 1025. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 Talks, Who’s Listening? Antioxid. Redox Signal. 2010, 13, 1649–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Wakabayashi, N.; Misra, V.; Biswal, S.; Lee, G.H.; Agoston, E.S.; Yamamoto, M.; Kensler, T.W. NRF2 Modulates Aryl Hydrocarbon Receptor Signaling: Influence on Adipogenesis. Mol. Cell. Biol. 2007, 27, 7188–7197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Shui, X.; Su, W.; He, Y.; Lu, X.; Zhang, Y.; Yan, G.; Huang, S.; Lei, W.; Chen, C. Baicalin inhibits inflammation and attenuates myocardial ischaemic injury by aryl hydrocarbon receptor. J. Pharm. Pharmacol. 2015, 67, 1756–1764. [Google Scholar] [CrossRef] [PubMed]
- Silva-Palacios, A.; Ostolga-Chavarría, M.; Sánchez-Garibay, C.; Rojas-Morales, P.; Galván-Arzate, S.; Buelna-Chontal, M.; Pavón, N.; Pedraza-Chaverrí, J.; Königsberg, M.; Zazueta, C. Sulforaphane protects from myocardial ischemia-reperfusion damage through the balanced activation of Nrf2/AhR. Free Radic. Biol. Med. 2019, 143, 331–340. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.; et al. RXRα Inhibits the NRF2-ARE Signaling Pathway through a Direct Interaction with the Neh7 Domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef] [Green Version]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [Green Version]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef]
- Zhang, J.; Ohta, T.; Maruyama, A.; Hosoya, T.; Nishikawa, K.; Maher, J.M.; Shibahara, S.; Itoh, K.; Yamamoto, M. BRG1 Interacts with Nrf2 to Selectively Mediate HO-1 Induction in Response to Oxidative Stress. Mol. Cell. Biol. 2006, 26, 7942–7952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, G.; Hebbar, V.; Nair, S.; Xu, C.; Li, W.; Lin, W.; Keum, Y.-S.; Han, J.; Gallo, M.A.; Kong, A.-N.T. Regulation of Nrf2 Transactivation Domain Activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem. 2004, 279, 23052–23060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Hosoya, T.; Maruyama, A.; Nishikawa, K.; Maher, J.M.; Ohta, T.; Motohashi, H.; Fukamizu, A.; Shibahara, S.; Itoh, K.; et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 2007, 404, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yu, S.; Kong, A.-N.T. Nrf2 Possesses a Redox-sensitive Nuclear Exporting Signal in the Neh5 Transactivation Domain. J. Biol. Chem. 2006, 281, 27251–27263. [Google Scholar] [CrossRef] [Green Version]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef] [Green Version]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated Turnover of Nrf2 Is Determined by at Least Two Separate Protein Domains, the Redox-sensitive Neh2 Degron and the Redox-insensitive Neh6 Degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Chin, Y.E.; Zhang, D.D. Acetylation of Nrf2 by p300/CBP Augments Promoter-Specific DNA Binding of Nrf2 during the Antioxidant Response. Mol. Cell. Biol. 2009, 29, 2658–2672. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yu, S.; Liu, T.; Kim, J.-H.; Blank, V.; Li, H.; Kong, A.-N.T. Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim. Biophys. Acta 2008, 1783, 1847–1856. [Google Scholar] [CrossRef] [Green Version]
- Nioi, P.; Nguyen, T.; Sherratt, P.J.; Pickett, C.B. The Carboxy-Terminal Neh3 Domain of Nrf2 Is Required for Transcriptional Activation. Mol. Cell. Biol. 2005, 25, 10895–10906. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, A.; Kang, M.-I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [Green Version]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J. Dimerization of Substrate Adaptors Can Facilitate Cullin-mediated Ubiquitylation of Proteins by a “Tethering” Mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Lo, S.-C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaspar, J.W.; Jaiswal, A.K. An Autoregulatory Loop between Nrf2 and Cul3-Rbx1 Controls Their Cellular Abundance. J. Biol. Chem. 2010, 285, 21349–21358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/-TrCP Promotes Glycogen Synthase Kinase 3-Dependent Degradation of the Nrf2 Transcription Factor in a Keap1-Independent Manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wang, Y.; Zhang, C.; Li, J.; Meng, Y.; Dou, M.; Noguchi, C.T.; Di, L. Inhibiting Glycogen Synthase Kinase 3 Reverses Obesity-Induced White Adipose Tissue Inflammation by Regulating Apoptosis Inhibitor of Macrophage/CD5L-Mediated Macrophage Migration. Arter. Thromb. Vasc. Biol. 2018, 38, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- Eldar-Finkelman, H.; Schreyer, S.A.; Shinohara, M.M.; LeBoeuf, R.C.; Krebs, E.G. Increased glycogen synthase kinase-3 activity in diabetes- and obesity-prone C57BL/6J mice. Diabetes 1999, 48, 1662–1666. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, X.; Han, L. Apoptosis of cardiomyocytes in diabetic cardiomyopathy involves overexpression of glycogen synthase kinase-3β. Biosci. Rep. 2019, 39, BSR20171307. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological Significance of Reactive Cysteine Residues of Keap1 in Determining Nrf2 Activity. Mol. Cell. Biol. 2008, 28, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Takaya, K.; Suzuki, T.; Motohashi, H.; Onodera, K.; Satomi, S.; Kensler, T.W.; Yamamoto, M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012, 53, 817–827. [Google Scholar] [CrossRef] [Green Version]
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.D.; Hannink, M. Distinct Cysteine Residues in Keap1 Are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodore, M.; Kawai, Y.; Yang, J.; Kleshchenko, Y.; Reddy, S.P.; Villalta, F.; Arinze, I.J. Multiple Nuclear Localization Signals Function in the Nuclear Import of the Transcription Factor Nrf2. J. Biol. Chem. 2008, 283, 8984–8994, Erratum in J. Biol. Chem. 2008, 283, 14176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuoka, F.; Motohashi, H.; Ishii, T.; Aburatani, H.; Engel, J.D.; Yamamoto, M. Genetic Evidence that Small Maf Proteins Are Essential for the Activation of Antioxidant Response Element-Dependent Genes. Mol. Cell. Biol. 2005, 25, 8044–8051. [Google Scholar] [CrossRef] [Green Version]
- Bloom, D.A.; Jaiswal, A.K. Phosphorylation of Nrf2 at Ser40 by Protein Kinase C in Response to Antioxidants Leads to the Release of Nrf2 from INrf2, but Is Not Required for Nrf2 Stabilization/Accumulation in the Nucleus and Transcriptional Activation of Antioxidant Response Element-mediated NAD(P)H:Quinone Oxidoreductase-1 Gene Expression. J. Biol. Chem. 2003, 278, 44675–44682. [Google Scholar] [CrossRef] [Green Version]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef]
- Jang, D.E.; Song, J.; Park, J.-W.; Yoon, S.-H.; Bae, Y.-S. Protein kinase CK2 activates Nrf2 via autophagic degradation of Keap1 and activation of AMPK in human cancer cells. BMB Rep. 2020, 53, 272–277. [Google Scholar] [CrossRef]
- Joo, M.S.; Kim, W.D.; Lee, K.Y.; Kim, J.H.; Koo, J.H.; Kim, S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016, 36, 1931–1942. [Google Scholar] [CrossRef] [Green Version]
- Matzinger, M.; Fischhuber, K.; Pölöske, D.; Mechtler, K.; Heiss, E.H. AMPK leads to phosphorylation of the transcription factor Nrf2, tuning transactivation of selected target genes. Redox Biol. 2020, 29, 101393. [Google Scholar] [CrossRef]
- Volz, N.; Boettler, U.; Winkler, S.; Teller, N.; Schwarz, C.; Bakuradze, T.; Eisenbrand, G.; Haupt, L.M.; Griffiths, L.; Stiebitz, H.; et al. Effect of Coffee Combining Green Coffee Bean Constituents with Typical Roasting Products on the Nrf2/ARE Pathway in Vitro and in Vivo. J. Agric. Food Chem. 2012, 60, 9631–9641. [Google Scholar] [CrossRef]
- Boettler, U.; Volz, N.; Teller, N.; Haupt, L.M.; Bakuradze, T.; Eisenbrand, G.; Bytof, G.; Lantz, I.; Griffiths, L.R.; Marko, D. Induction of antioxidative Nrf2 gene transcription by coffee in humans: Depending on genotype? Mol. Biol. Rep. 2012, 39, 7155–7162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerotic, D.; Matic, M.; Suvakov, S.; Vucicevic, K.; Damjanovic, T.; Savic-Radojevic, A.; Pljesa-Ercegovac, M.; Coric, V.; Stefanovic, A.; Ivanisevic, J.; et al. Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients. Toxins 2019, 11, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, K.N.; Florentino, R.M.; França, A.; Filho, A.C.M.L.; Dos Santos, M.L.; Missiaggia, D.; Fonseca, M.D.C.; Costa, I.B.; Vidigal, P.V.T.; Nathanson, M.H.; et al. Polymorphism in the Promoter Region of NFE2L2 Gene Is a Genetic Marker of Susceptibility to Cirrhosis Associated with Alcohol Abuse. Int. J. Mol. Sci. 2019, 20, 3589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korytina, G.F.; Akhmadishina, L.Z.; Aznabaeva, Y.; Kochetova, O.V.; Zagidullin, N.; Kzhyshkowska, J.G.; Zagidullin, S.Z.; Viktorova, T.V. Associations of the NRF2/KEAP1 pathway and antioxidant defense gene polymorphisms with chronic obstructive pulmonary disease. Gene 2019, 692, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Siedlinski, M.; Postma, D.S.; Boer, J.M.; van der Steege, G.; Schouten, J.P.; Smit, H.A.; Boezen, H.M. Level and course of FEV1 in relation to polymorphisms in NFE2L2 and KEAP1 in the general population. Respir. Res. 2009, 10, 73. [Google Scholar] [CrossRef] [Green Version]
- Testa, A.; Leonardis, D.; Spoto, B.; Sanguedolce, M.C.; Parlongo, R.M.; Pisano, A.; Tripepi, G.; Mallamaci, F.; Zoccali, C. A polymorphism in a major antioxidant gene (Kelch-like ECH-associated protein 1) predicts incident cardiovascular events in chronic kidney disease patients: An exploratory study. J. Hypertens. 2016, 34, 928–934. [Google Scholar] [CrossRef]
- Marzec, J.M.; Christie, J.D.; Reddy, S.P.; Jedlicka, A.E.; Vuong, H.; Lanken, P.N.; Aplenc, R.; Yamamoto, T.; Yamamoto, M.; Cho, H.-Y.; et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007, 21, 2237–2246. [Google Scholar] [CrossRef]
- Figarska, S.M.; Vonk, J.M.; Boezen, H.M. NFE2L2 polymorphisms, mortality, and metabolism in the general population. Physiol. Genom. 2014, 46, 411–417. [Google Scholar] [CrossRef]
- Scutt, G.; Overall, A.; Bakrania, P.; Krasteva, E.; Parekh, N.; Ali, K.; Davies, J.G.; Rajkumar, C. The Association of a Single-Nucleotide Polymorphism in the Nuclear Factor (Erythroid-Derived 2)-Like 2 Gene With Adverse Drug Reactions, Multimorbidity, and Frailty in Older People. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 1050–1057. [Google Scholar] [CrossRef]
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in obesity, diabetes, and related disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef]
- Ozata, M.; Mergen, M.; Oktenli, C.; Aydin, A.; Sanisoglu, S.Y.; Bolu, E.; Yilmaz, M.; Sayal, A.; Isimer, A.; Ozdemir, I. Increased oxidative stress and hypozincemia in male obesity. Clin. Biochem. 2002, 35, 627–631. [Google Scholar] [CrossRef]
- Adnan, T.; Amin, M.N.; Uddin, G.; Hussain, S.; Sarwar, S.; Hossain, K.; Uddin, S.N.; Islam, M.S. Increased concentration of serum MDA, decreased antioxidants and altered trace elements and macro-minerals are linked to obesity among Bangladeshi population. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Monzo-Beltran, L.; Vazquez-Tarragón, A.; Cerdà, C.; Garcia-Perez, P.; Iradi, A.; Sánchez, C.; Climent, B.; Tormos, C.; Vázquez-Prado, A.; Girbés, J.; et al. One-year follow-up of clinical, metabolic and oxidative stress profile of morbid obese patients after laparoscopic sleeve gastrectomy. 8-oxo-dG as a clinical marker. Redox Biol. 2017, 12, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, A.; Korac, A.; Galic, B.S.; Buzadzic, B.; Otasevic, V.; Stancic, A.; Vucetic, M.; Markelić, M.; Velickovic, K.; Golic, I.; et al. Differences in the redox status of human visceral and subcutaneous adipose tissues—Relationships to obesity and metabolic risk. Metabolism 2014, 63, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Morya, R.K.; Saha, S.; Singh, P.K.; Bhadada, S.K.; Rana, S.V. Oxidative stress and inflammatory markers in type 2 diabetic patients. Eur. J. Clin. Investig. 2020, 50, e13238. [Google Scholar] [CrossRef]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T.; Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced Glycation End Products and Oxidative Stress in Type 2 Diabetes Mellitus. Biomolecules 2015, 5, 194–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhu, C.; Liu, T.; Zhang, W.; Liu, X.; Li, P.; Zhu, T. Epigallocatechin-3-gallate ameliorates glucolipid metabolism and oxidative stress in type 2 diabetic rats. Diabetes Vasc. Dis. Res. 2020, 17, 1479164120966998. [Google Scholar] [CrossRef]
- Wang, W.; He, Y.; Liu, Q. Parthenolide plays a protective role in the liver of mice with metabolic dysfunction-associated fatty liver disease through the activation of the HIPPO pathway. Mol. Med. Rep. 2021, 24, 487. [Google Scholar] [CrossRef]
- Perdomo, L.; Beneit, N.; Otero, Y.F.; Escribano, Ó.; Díaz-Castroverde, S.; Gómez-Hernández, A.; Benito, M. Protective role of oleic acid against cardiovascular insulin resistance and in the early and late cellular atherosclerotic process. Cardiovasc. Diabetol. 2015, 14, 75. [Google Scholar] [CrossRef] [Green Version]
- Zurbau, A.; Duvnjak, L.S.; Magas, S.; Jovanovski, E.; Miocic, J.; Jenkins, A.L.; Jenkins, D.J.A.; Josse, R.G.; Leiter, L.A.; Sievenpiper, J.L.; et al. Co-administration of viscous fiber, Salba-chia and ginseng on glycemic management in type 2 diabetes: A double-blind randomized controlled trial. Eur. J. Nutr. 2021, 60, 3071–3083. [Google Scholar] [CrossRef]
- Ren, B.C.; Zhang, Y.F.; Liu, S.S.; Cheng, X.J.; Yang, X.; Cui, X.G.; Zhao, X.R.; Zhao, H.; Hao, M.F.; Li, M.D.; et al. Curcumin alleviates oxidative stress and inhibits apoptosis in diabetic cardiomyopathy via Sirt1-Foxo1 and PI3K-Akt signalling pathways. J. Cell. Mol. Med. 2020, 24, 12355–12367. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, Y.; Li, X.; Liu, J.; Wang, J.; Lu, Y. Sulforaphane Regulates Glucose and Lipid Metabolisms in Obese Mice by Restraining JNK and Activating Insulin and FGF21 Signal Pathways. J. Agric. Food Chem. 2021, 69, 13066–13079. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gao, J.; Ke, W.; Wang, J.; Li, D.; Liu, R.; Jia, Y.; Wang, X.; Chen, X.; Chen, F.; et al. Resveratrol reduces obesity in high-fat diet-fed mice via modulating the composition and metabolic function of the gut microbiota. Free Radic. Biol. Med. 2020, 156, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wang, S.; Zhang, H.; Yang, G.; Bai, Y.; Liu, P.; Meng, L.; Jiang, X.; Xin, Y. Sulforaphane prevents angiotensin II-induced cardiomyopathy by activation of Nrf2 through epigenetic modification. J. Cell. Mol. Med. 2021, 25, 4408–4419. [Google Scholar] [CrossRef] [PubMed]
- Darband, S.G.; Sadighparvar, S.; Yousefi, B.; Kaviani, M.; Ghaderi-Pakdel, F.; Mihanfar, A.; Rahimi, Y.; Mobaraki, K.; Majidinia, M. Quercetin attenuated oxidative DNA damage through NRF2 signaling pathway in rats with DMH induced colon carcinogenesis. Life Sci. 2020, 253, 117584. [Google Scholar] [CrossRef]
- Zhao, P.; Hu, Z.; Ma, W.; Zang, L.; Tian, Z.; Hou, Q. Quercetin alleviates hyperthyroidism-induced liver damage via Nrf2 Signaling pathway. BioFactors 2020, 46, 608–619. [Google Scholar] [CrossRef]
- Tan, Y.; Ichikawa, T.; Li, J.; Si, Q.; Yang, H.; Chen, X.; Goldblatt, C.S.; Meyer, C.J.; Li, X.; Cai, L.; et al. Diabetic Downregulation of Nrf2 Activity via ERK Contributes to Oxidative Stress–Induced Insulin Resistance in Cardiac Cells In Vitro and In Vivo. Diabetes 2011, 60, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Uruno, A.; Yagishita, Y.; Yamamoto, M. The Keap1–Nrf2 system and diabetes mellitus. Arch. Biochem. Biophys. 2015, 566, 76–84. [Google Scholar] [CrossRef] [Green Version]
- Adam, M.; Imboden, M.; Schaffner, E.; Boes, E.; Kronenberg, F.; Pons, M.; Bettschart, R.; Barthelemy, J.-C.; Schindler, C.; Probst-Hensch, N. The adverse impact of obesity on heart rate variability is modified by a NFE2L2 gene variant: The SAPALDIA cohort. Int. J. Cardiol. 2017, 228, 341–346. [Google Scholar] [CrossRef] [Green Version]
- Gusti, A.; Qusti, S.; Alshammari, E.; Toraih, E.; Fawzy, M. Antioxidants-Related Superoxide Dismutase (SOD), Catalase (CAT), Glutathione Peroxidase (GPX), Glutathione-S-Transferase (GST), and Nitric Oxide Synthase (NOS) Gene Variants Analysis in an Obese Population: A Preliminary Case-Control Study. Antioxidants 2021, 10, 595. [Google Scholar] [CrossRef]
- Montano, M.A.E.; Lera, J.P.B.; Gottlieb, M.; Schwanke, C.; Da Rocha, M.I.U.M.; Manica-Cattani, M.F.; Dos Santos, G.F.; da Cruz, I. Association between manganese superoxide dismutase (MnSOD) gene polymorphism and elderly obesity. Mol. Cell. Biochem. 2009, 328, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Duarte, T.; da Cruz, I.; Barbisan, F.; Capelleto, D.; Moresco, R.; Duarte, M.M.M.F. The effects of rosuvastatin on lipid-lowering, inflammatory, antioxidant and fibrinolytics blood biomarkers are influenced by Val16Ala superoxide dismutase manganese-dependent gene polymorphism. Pharm. J. 2016, 16, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, M.G.V.; Schwanke, C.H.A.; Santos, A.F.R.; Jobim, P.F.; Müssel, D.P.; da Cruz, I. Association among oxidized LDL levels, MnSOD, apolipoprotein E polymorphisms, and cardiovascular risk factors in a south Brazilian region population. Genet. Mol. Res. 2005, 4, 691–703. [Google Scholar] [PubMed]
- Nakanishi, S.; Yamane, K.; Ohishi, W.; Nakashima, R.; Yoneda, M.; Nojima, H.; Watanabe, H.; Kohno, N. Manganese superoxide dismutase Ala16Val polymorphism is associated with the development of type 2 diabetes in Japanese-Americans. Diabetes Res. Clin. Pract. 2008, 81, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, T.; Tanaka, Y.; Piao, L.; Nagasaka, K.; Sakai, K.; Ogihara, T.; Nakajima, K.; Watada, H.; Kawamori, R. The polymorphism of manganese superoxide dismutase is associated with diabetic nephropathy in Japanese type 2 diabetic patients. J. Hum. Genet. 2003, 48, 0138–0141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollsten, A.; Marklund, S.L.; Wessman, M.; Svensson, M.; Forsblom, C.; Parkkonen, M.; Brismar, K.; Groop, P.-H.; Dahlquist, G. A Functional Polymorphism in the Manganese Superoxide Dismutase Gene and Diabetic Nephropathy. Diabetes 2007, 56, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollsten, A.; Jorsal, A.; Lajer, M.; Vionnet, N.; Tarnow, L. The V16A polymorphism in SOD2 is associated with increased risk of diabetic nephropathy and cardiovascular disease in type 1 diabetes. Diabetologia 2009, 52, 2590–2593. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Sun, J.; Chang, X.; Wang, J.; Luo, M.; Wintergerst, K.A.; Miao, L.; Cai, L. Genetic variants of nuclear factor erythroid-derived 2-like 2 associated with the complications in Han descents with type 2 diabetes mellitus of Northeast China. J. Cell. Mol. Med. 2016, 20, 2078–2088. [Google Scholar] [CrossRef] [Green Version]
- Hernández Guerrero, C.; Hernández Chávez, P.; Martínez Castro, N.; Parra Carriedo, A.; García Del Rio, S.; Pérez Lizaur, A. Glutathione peroxidase-1 PRO200LEU polymorphism (Rs1050450) is associated with morbid obesity independently of the presence of prediabetes or diabetes in women from central Mexico. Nutr. Hosp. 2015, 32, 1516–1525. [Google Scholar] [CrossRef]
- Buraczynska, M.; Buraczynska, K.; Dragan, M.; Ksiazek, A. Pro198Leu Polymorphism in the Glutathione Peroxidase 1 Gene Contributes to Diabetic Peripheral Neuropathy in Type 2 Diabetes Patients. Neuromolecular Med. 2017, 19, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.; Prior, S.; Li, K.; Ireland, H.; Bain, S.; Hurel, S.; Cooper, J.; Humphries, S.; Stephens, J. Association between the rs1050450 glutathione peroxidase-1 (C > T) gene variant and peripheral neuropathy in two independent samples of subjects with diabetes mellitus. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 417–425. [Google Scholar] [CrossRef]
- Liu, D.; Liu, L.; Hu, Z.; Song, Z.; Wang, Y.; Chen, Z. Evaluation of the oxidative stress–related genes ALOX5, ALOX5AP, GPX1, GPX3 and MPO for contribution to the risk of type 2 diabetes mellitus in the Han Chinese population. Diabetes Vasc. Dis. Res. 2018, 15, 336–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Osorio, A.S.; González-Reyes, S.; García-Niño, W.R.; Moreno-Macías, H.; Arellano, M.E.R.; Vargas-Alarcón, G.; Zuñiga, J.; Barquera, R.; Pedraza-Chaverri, J. Association of Nuclear Factor-Erythroid 2-Related Factor 2, Thioredoxin Interacting Protein, and Heme Oxygenase-1 Gene Polymorphisms with Diabetes and Obesity in Mexican Patients. Oxidative Med. Cell. Longev. 2016, 2016, 7367641, Erratum in Oxidative Med. Cell. Longev. 2017, 2017, 7543194. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, M.; Chen, J.; Zhu, L.; Liu, J.; Xu, J. Association of Nuclear Factor Erythroid-2-Related Actor 2 Gene Polymorphisms with Diabetic Nephropathy in Chinese Patients. Int. J. Gen. Med. 2021, 14, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, H.; Liu, J.; Ouyang, Y.; Wang, D.; Bao, W.; Liu, L. Association between the NF-E2 Related Factor 2 Gene Polymorphism and Oxidative Stress, Anti-Oxidative Status, and Newly-Diagnosed Type 2 Diabetes Mellitus in a Chinese Population. Int. J. Mol. Sci. 2015, 16, 16483–16496. [Google Scholar] [CrossRef] [Green Version]
- Teena, R.; Dhamodharan, U.; Ali, D.; Rajesh, K.; Ramkumar, K.M. Genetic Polymorphism of the Nrf2 Promoter Region (rs35652124) Is Associated with the Risk of Diabetic Foot Ulcers. Oxidative Med. Cell. Longev. 2020, 2020, 9825028. [Google Scholar] [CrossRef]
- Teena, R.; Dhamodharan, U.; Jayasuriya, R.; Ali, D.; Kesavan, R.; Ramkumar, K.M. Analysis of the Exonic Single Nucleotide Polymorphism rs182428269 of the NRF2 Gene in Patients with Diabetic Foot Ulcer. Arch. Med. Res. 2021, 52, 224–232. [Google Scholar] [CrossRef]
- Shin, J.H.; Lee, K.-M.; Shin, J.; Kang, K.D.; Nho, C.W.; Cho, Y.S. Genetic risk score combining six genetic variants associated with the cellular NRF2 expression levels correlates with Type 2 diabetes in the human population. Genes Genom. 2019, 41, 537–545. [Google Scholar] [CrossRef]
- Da Costa, R.M.; Rodrigues, D.; Pereira, C.A.; Silva, J.; Alves, J.V.; Lobato, N.S.; Tostes, R.C. Nrf2 as a Potential Mediator of Cardiovascular Risk in Metabolic Diseases. Front. Pharmacol. 2019, 10, 382. [Google Scholar] [CrossRef] [Green Version]
- Howden, R. Nrf2and Cardiovascular Defense. Oxidative Med. Cell. Longev. 2013, 2013, 104308. [Google Scholar] [CrossRef] [Green Version]
- Buelna-Chontal, M.; Zazueta, C. Redox activation of Nrf2 & NF-κB: A double end sword? Cell. Signal. 2013, 25, 2548–2557. [Google Scholar] [CrossRef]
- Tebay, L.E.; Robertson, H.; Durant, S.T.; Vitale, S.R.; Penning, T.M.; Dinkova-Kostova, A.T.; Hayes, J.D. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free. Radic. Biol. Med. 2015, 88, 108–146. [Google Scholar] [CrossRef] [Green Version]
- Shimoyama, Y.; Mitsuda, Y.; Tsuruta, Y.; Hamajima, N.; Niwa, T. Polymorphism of Nrf2, an Antioxidative Gene, is Associated with Blood Pressure and Cardiovascular Mortality in Hemodialysis Patients. Int. J. Med. Sci. 2014, 11, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barajas, B.; Che, N.; Yin, F.; Rowshanrad, A.; Orozco, L.D.; Gong, K.W.; Wang, X.; Castellani, L.W.; Reue, K.; Lusis, A.J.; et al. NF-E2–Related Factor 2 Promotes Atherosclerosis by Effects on Plasma Lipoproteins and Cholesterol Transport That Overshadow Antioxidant Protection. Arter. Thromb. Vasc. Biol. 2011, 31, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freigang, S.; Ampenberger, F.; Spohn, G.; Heer, S.; Shamshiev, A.T.; Kisielow, J.; Hersberger, M.; Yamamoto, M.; Bachmann, M.F.; Kopf, M. Nrf2 is essential for cholesterol crystal-induced inflammasome activation and exacerbation of atherosclerosis. Eur. J. Immunol. 2011, 41, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Ruotsalainen, A.-K.; Lappalainen, J.P.; Heiskanen, E.; Merentie, M.; Sihvola, V.; Näpänkangas, J.; Lottonen-Raikaslehto, L.; Kansanen, E.; Adinolfi, S.; Kaarniranta, K.; et al. Nuclear factor E2-related factor 2 deficiency impairs atherosclerotic lesion development but promotes features of plaque instability in hypercholesterolaemic mice. Cardiovasc. Res. 2018, 115, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Sarutipaiboon, I.; Settasatian, N.; Komanasin, N.; Kukongwiriyapan, U.; Sawanyawisuth, K.; Intharaphet, P.; Senthong, V.; Settasatian, C. Association of Genetic Variations in NRF2, NQO1, HMOX1, and MT with Severity of Coronary Artery Disease and Related Risk Factors. Cardiovasc. Toxicol. 2020, 20, 176–189. [Google Scholar] [CrossRef]
- Miyazawa, M.; Tsuji, Y. Evidence for a novel antioxidant function and isoform-specific regulation of the human p66Shc gene. Mol. Biol. Cell 2014, 25, 2116–2127. [Google Scholar] [CrossRef]
- Napoli, C.; Martin-Padura, I.; de Nigris, F.; Giorgio, M.; Mansueto, G.; Somma, P.; Condorelli, M.; Sica, G.; de Rosa, G.; Pelicci, P. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc. Natl. Acad. Sci. USA 2003, 100, 2112–2116. [Google Scholar] [CrossRef] [Green Version]
- Sentinelli, F.; Romeo, S.; Barbetti, F.; Berni, A.; Filippi, E.; Fanelli, M.; Fallarino, M.; Baroni, M.G. Search for genetic variants in the p66Shc longevity gene by PCR-single strand conformational polymorphism in patients with early-onset cardiovascular disease. BMC Genet. 2006, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Campbell, M.R.; Lacher, S.E.; Cho, H.-Y.; Wan, M.; Crowl, C.L.; Chorley, B.N.; Bond, G.L.; Kleeberger, S.R.; Slattery, M.; et al. A Polymorphic Antioxidant Response Element Links NRF2/sMAF Binding to Enhanced MAPT Expression and Reduced Risk of Parkinsonian Disorders. Cell Rep. 2016, 15, 830–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linna-Kuosmanen, S.; Viitala, S.; Laitinen, T.; Peräkylä, M.; Pölönen, P.; Kansanen, E.; Leinonen, H.; Raju, S.; Wienecke-Baldacchino, A.; Närvänen, A.; et al. The Effects of Sequence Variation on Genome-wide NRF2 Binding—New Target Genes and Regulatory SNPs. Nucleic Acids Res. 2016, 44, 1760–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, H.; Matsumoto, M.; Shindo, T.; Saigusa, D.; Kato, H.; Suzuki, K.; Sato, M.; Ishii, Y.; Shimokawa, H.; Igarashi, K. Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J. Biol. Chem. 2020, 295, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Mannami, T.; Iwai, N. Association of a promoter variant of the haeme oxygenase-1 gene with hypertension in women. J. Hypertens. 2003, 21, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Han, S.J.; Kang, E.S.; Kim, H.J.; Kim, S.H.; Chun, S.W.; Ahn, C.W.; Cha, B.S.; Nam, M.; Lee, H.C. The C609T variant of NQO1 is associated with carotid artery plaques in patients with type 2 diabetes. Mol. Genet. Metab. 2009, 97, 85–90. [Google Scholar] [CrossRef]
- Martin, N.J.; Collier, A.; Bowen, L.D.; Pritsos, K.L.; Goodrich, G.G.; Arger, K.; Cutter, G.; Pritsos, C.A. Polymorphisms in the NQO1, GSTT and GSTM genes are associated with coronary heart disease and biomarkers of oxidative stress. Mutat. Res. 2009, 674, 93–100. [Google Scholar] [CrossRef]
- Yu, H.; Liu, H.; Wang, L.-E.; Wei, Q. A Functional NQO1 609C>T Polymorphism and Risk of Gastrointestinal Cancers: A Meta-Analysis. PLoS ONE 2012, 7, e30566. [Google Scholar] [CrossRef] [Green Version]
- Duarte, M.M.; Moresco, R.N.; Duarte, T.; Santi, A.; Bagatini, M.D.; Da Cruz, I.B.; Schetinger, M.R.; Loro, V.L. Oxidative stress in hypercholesterolemia and its association with Ala16Val superoxide dismutase gene polymorphism. Clin. Biochem. 2010, 43, 1118–1123. [Google Scholar] [CrossRef]
- Lopes, R.A.; Neves, K.B.; Tostes, R.C.; Montezano, A.C.; Touyz, R.M. Downregulation of Nuclear Factor Erythroid 2–Related Factor and Associated Antioxidant Genes Contributes to Redox-Sensitive Vascular Dysfunction in Hypertension. Hypertension 2015, 66, 1240–1250. [Google Scholar] [CrossRef] [Green Version]
- Kunnas, T.; Määttä, K.; Nikkari, S.T. Genetic Polymorphisms of Transcription Factor NRF2 and of its Host Gene Sulfiredoxin (SRXN1) are Associated with Cerebrovascular Disease in a Finnish Cohort, the TAMRISK Study. Int. J. Med. Sci. 2016, 13, 325–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marczak, E.D.; Marzec, J.; Zeldin, D.; Kleeberger, S.; Brown, N.J.; Pretorius, M.; Lee, C.R. Polymorphisms in the transcription factor NRF2 and forearm vasodilator responses in humans. Pharm. Genom. 2012, 22, 620–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.-L.; Sun, L.; Wang, Y.-X.; Sun, B.-H.; Li, Y.-F.; Jin, Y.-L. Association between HO-1 gene promoter polymorphisms and diseases (Review). Mol. Med. Rep. 2022, 25, 29. [Google Scholar] [CrossRef] [PubMed]
- Mansego, M.L.; Solar, G.D.M.; Alonso, M.P.; Martínez, F.; Sáez, G.T.; Escudero, J.C.M.; Redón, J.; Chaves, F.J. Polymorphisms of antioxidant enzymes, blood pressure and risk of hypertension. J. Hypertens. 2011, 29, 492–500. [Google Scholar] [CrossRef]
- An, S.H.; Chang, B.C.; Lee, K.E.; Gwak, H.S. Influence of UDP-Glucuronosyltransferase Polymorphisms on Stable Warfarin Doses in Patients with Mechanical Cardiac Valves. Cardiovasc. Ther. 2015, 33, 324–328. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Nakajima, M.; Higashi, E.; Yamanaka, H.; Yokoi, T. Genetic polymorphisms in the 5′-flanking region of human UDP-glucuronosyltransferase 2B7 affect the Nrf2-dependent transcriptional regulation. Pharm. Genom. 2008, 18, 709–720. [Google Scholar] [CrossRef]
- Pereira, N.L.; Rihal, C.S.; So, D.Y.; Rosenberg, Y.; Lennon, R.J.; Mathew, V.; Goodman, S.G.; Weinshilboum, R.M.; Wang, L.; Baudhuin, L.M.; et al. Clopidogrel Pharmacogenetics. Circ. Cardiovasc. Interv. 2019, 12, e007811. [Google Scholar] [CrossRef]
- Pare, G.; Ross, S.; Mehta, S.R.; Yusuf, S.; Anand, S.S.; Connolly, S.J.; Fox, K.; Eikelboom, J.W. Effect of PON1 Q192R Genetic Polymorphism on Clopidogrel Efficacy and Cardiovascular Events in the Clopidogrel in the Unstable Angina to Prevent Recurrent Events Trial and the Atrial Fibrillation Clopidogrel Trial With Irbesartan for Prevention of Vascular Events. Circ. Cardiovasc. Genet. 2012, 5, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Kosmidou, M.; Hatzitolios, A.I.; Molyva, D.; Raikos, N.; Savopoulos, C.; Daferera, N.; Kokkas, V.; Goulas, A. An association study between catalase -262C>T gene polymorphism, sodium-lithium countertrasport activity, insulin resistance, blood lipid parameters and their response to atorvastatin, in Greek dyslipidaemic patients and normolipidaemic controls. Free Radic. Res. 2009, 43, 385–389. [Google Scholar] [CrossRef]
- Watanabe, L.M.; Bueno, A.C.; de Lima, L.F.; Ferraz-Bannitz, R.; Dessordi, R.; Guimarães, M.P.; Foss-Freitas, M.C.; Barbosa, F., Jr.; Navarro, A.M. Genetically determined variations of selenoprotein P are associated with antioxidant, muscular, and lipid biomarkers in response to Brazil nut consumption by patients using statins. Br. J. Nutr. 2021, 127, 679–686. [Google Scholar] [CrossRef]
- Van Schaik, L.; Kettle, C.; Green, R.; Irving, H.R.; Rathner, J.A. Effects of Caffeine on Brown Adipose Tissue Thermogenesis and Metabolic Homeostasis: A Review. Front. Neurosci. 2021, 15, 621356. [Google Scholar] [CrossRef]
- Lara-Guzmán, O.J.; Álvarez, R.; Muñoz-Durango, K. Changes in the plasma lipidome of healthy subjects after coffee consumption reveal potential cardiovascular benefits: A randomized controlled trial. Free Radic. Biol. Med. 2021, 176, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Krittanawong, C.; Tunhasiriwet, A.; Wang, Z.; Farrell, A.M.; Chirapongsathorn, S.; Zhang, H.; Kitai, T.; Mehta, D. Is caffeine or coffee consumption a risk for new-onset atrial fibrillation? A systematic review and meta-analysis. Eur. J. Prev. Cardiol. 2021, 28, e13–e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolb, H.; Martin, S.; Kempf, K. Coffee and Lower Risk of Type 2 Diabetes: Arguments for a Causal Relationship. Nutrients 2021, 13, 1144. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Liu, Y.; Hu, J.; Gao, Y.; Ma, Y.; Wen, D. Chlorogenic Acid-Induced Gut Microbiota Improves Metabolic Endotoxemia. Front. Endocrinol. 2021, 12, 762691. [Google Scholar] [CrossRef]
- Redondo-Puente, M.; Mateos, R.; Seguido, M.A.; García-Cordero, J.; González, S.; Tarradas, R.M.; Bravo-Clemente, L.; Sarriá, B. Appetite and Satiety Effects of the Acute and Regular Consumption of Green Coffee Phenols and Green Coffee Phenol/Oat β-Glucan Nutraceuticals in Subjects with Overweight and Obesity. Foods 2021, 10, 2511. [Google Scholar] [CrossRef]
- Mateos, R.; García-Cordero, J.; Bravo-Clemente, L.; Sarriá, B. Evaluation of novel nutraceuticals based on the combination of oat beta-glucans and a green coffee phenolic extract to combat obesity and its comorbidities. A randomized, dose–response, parallel trial. Food Funct. 2021, 13, 574–586. [Google Scholar] [CrossRef]
- Stevens, L.M.; Linstead, E.; Hall, J.L.; Kao, D.P. Association Between Coffee Intake and Incident Heart Failure Risk: A Machine Learning Analysis of the FHS, the ARIC Study, and the CHS. Circ. Heart Fail. 2021, 14, e006799. [Google Scholar] [CrossRef]
- Zhou, A.; Hyppönen, E. Habitual coffee intake and plasma lipid profile: Evidence from UK Biobank. Clin. Nutr. 2021, 40, 4404–4413. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, M.; Yuan, S.; Liu, X. Coffee consumption and risk of coronary artery disease. Eur. J. Prev. Cardiol. 2020, 29, e29–e31. [Google Scholar] [CrossRef]
- Nordestgaard, A.T. Causal relationship from coffee consumption to diseases and mortality: A review of observational and Mendelian randomization studies including cardiometabolic diseases, cancer, gallstones and other diseases. Eur. J. Nutr. 2021, 61, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Siller-Lopez, F.; Garzón-Castaño, S.; Ramos-Márquez, M.E.; Hernandez-Cañaveral, I. Association of Paraoxonase-1 Q192R (rs662) Single Nucleotide Variation with Cardiovascular Risk in Coffee Harvesters of Central Colombia. J. Toxicol. 2017, 2017, 6913106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayram, B.; Ozcelik, B.; Grimm, S.; Roeder, T.; Schrader, C.; Ernst, I.M.; Wagner, A.E.; Grune, T.; Frank, J.; Rimbach, G. A Diet Rich in Olive Oil Phenolics Reduces Oxidative Stress in the Heart of SAMP8 Mice by Induction of Nrf2-Dependent Gene Expression. Rejuvenation Res. 2012, 15, 71–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassmann, U.; Haupt, L.M.; Smith, R.A.; Winkler, S.; Bytof, G.; Lantz, I.; Griffiths, L.R.; Marko, D. Potential antioxidant response to coffee—A matter of genotype? Meta Gene 2014, 2, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yue, W.; Zhang, L.; Zhao, X.; Ma, L.; Yang, X.; Zhang, C.; Wang, Y.; Gu, M. Polymorphisms in GSTM1, CYP1A1, CYP2E1, and CYP2D6 are Associated with Susceptibility and Chemotherapy Response in Non-small-cell Lung Cancer Patients. Lung 2012, 190, 91–98. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.; Geller, F.; Prokopenko, I.; Feenstra, B.; Aben, K.K.; Franke, B.; den Heijer, M.; Kovacs, P.; Stumvoll, M.; et al. Sequence variants at CYP1A1–CYP1A2 and AHR associate with coffee consumption. Hum. Mol. Genet. 2011, 20, 2071–2077. [Google Scholar] [CrossRef]
- Amin, N.; Byrne, E.; Johnson, J.; Chenevix-Trench, G.; Walter, S.; Nolte, I.M.; Vink, J.; Rawal, R.; Mangino, M.; Teumer, A.; et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol. Psychiatry 2012, 17, 1116–1129. [Google Scholar] [CrossRef]
- McMahon, G.; Taylor, A.E.; Smith, G.D.; Munafò, M.R. Phenotype Refinement Strengthens the Association of AHR and CYP1A1 Genotype with Caffeine Consumption. PLoS ONE 2014, 9, e103448. [Google Scholar] [CrossRef] [Green Version]
- Nault, R.; Doskey, C.M.; Fader, K.A.; Rockwell, C.; Zacharewski, T.; Zacharewski, T.R. Comparison of Hepatic NRF2 and Aryl Hydrocarbon Receptor Binding in 2,3,7,8-Tetrachlorodibenzo-p-dioxin–Treated Mice Demonstrates NRF2-Independent PKM2 Induction. Mol. Pharmacol. 2018, 94, 876–884. [Google Scholar] [CrossRef] [Green Version]
- Van den Bogaard, E.H.; Bergboer, J.G.; Vonk-Bergers, M.; Van Vlijmen-Willems, I.M.; Hato, S.V.; Van Der Valk, P.G.; Schröder, J.M.; Joosten, I.; Zeeuwen, P.L.; Schalkwijk, J. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J. Clin. Investig. 2013, 123, 917–927. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Bao, R.-K.; Zhu, S.-Y.; Talukder, M.; Cui, J.-G.; Zhang, H.; Li, X.-N.; Li, J.-L. Lycopene prevents DEHP-induced hepatic oxidative stress damage by crosstalk between AHR–Nrf2 pathway. Environ. Pollut. 2021, 285, 117080. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Agrawal, S. A Global Perspective of Fine Particulate Matter Pollution and Its Health Effects. Rev. Environ. Contam. Toxicol. 2018, 244, 5–51. [Google Scholar] [CrossRef]
- Kwon, H.-S.; Ryu, M.H.; Carlsten, C. Ultrafine particles: Unique physicochemical properties relevant to health and disease. Exp. Mol. Med. 2020, 52, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Dubowsky, S.D.; Suh, H.; Schwartz, J.; Coull, B.A.; Gold, D.R. Diabetes, Obesity, and Hypertension May Enhance Associations between AirPollution and Markers of Systemic Inflammation. Environ. Health Perspect. 2006, 114, 992–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rückerl, R.; Hampel, R.; Breitner, S.; Cyrys, J.; Kraus, U.; Carter, J.; Dailey, L.; Devlin, R.B.; Diaz-Sanchez, D.; Koenig, W.; et al. Associations between ambient air pollution and blood markers of inflammation and coagulation/fibrinolysis in susceptible populations. Environ. Int. 2014, 70, 32–49. [Google Scholar] [CrossRef] [PubMed]
- Haberzettl, P.; Conklin, D.J.; Abplanalp, W.T.; Bhatnagar, A.; O’Toole, T.E. Inhalation of Fine Particulate Matter Impairs Endothelial Progenitor Cell Function Via Pulmonary Oxidative Stress. Arter. Thromb. Vasc. Biol. 2018, 38, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Ma, Y.; Luo, J.; Li, D.; Jiang, M.; Jiang, Q.; Pi, J.; Chen, R.; Chen, W.; Zhang, R.; et al. The Role of Nrf2 in the PM-Induced Vascular Injury Under Real Ambient Particulate Matter Exposure in C57/B6 Mice. Front. Pharmacol. 2021, 12, 618023. [Google Scholar] [CrossRef]
- Ge, C.; Hu, L.; Lou, D.; Li, Q.; Feng, J.; Wu, Y.; Tan, J.; Xu, M. Nrf2 deficiency aggravates PM2.5-induced cardiomyopathy by enhancing oxidative stress, fibrosis and inflammation via RIPK3-regulated mitochondrial disorder. Aging 2020, 12, 4836–4865. [Google Scholar] [CrossRef]
- Canova, C.; Dunster, C.; Kelly, F.J.; Minelli, C.; Shah, P.L.; Caneja, C.; Tumilty, M.K.; Burney, P. PM10-induced Hospital Admissions for Asthma and Chronic Obstructive Pulmonary Disease: The modifying effect of individual characteristics. Epidemiology 2012, 23, 607–615. [Google Scholar] [CrossRef]
- Wittkopp, S.; Staimer, N.; Tjoa, T.; Stinchcombe, T.; Daher, N.; Schauer, J.J.; Shafer, M.M.; Sioutas, C.; Gillen, D.L.; Delfino, R.J. Nrf2-related gene expression and exposure to traffic-related air pollution in elderly subjects with cardiovascular disease: An exploratory panel study. J. Expo. Sci. Environ. Epidemiol. 2016, 26, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Hou, W.; Hu, S.; Li, C.; Ma, H.; Wang, Q.; Meng, G.; Guo, T.; Zhang, J. Cigarette Smoke Induced Lung Barrier Dysfunction, EMT, and Tissue Remodeling: A Possible Link between COPD and Lung Cancer. BioMed Res. Int. 2019, 2019, 2025636. [Google Scholar] [CrossRef] [PubMed]
- Giebe, S.; Hofmann, A.; Brux, M.; Lowe, F.; Breheny, D.; Morawietz, H.; Brunssen, C. Comparative study of the effects of cigarette smoke versus next generation tobacco and nicotine product extracts on endothelial function. Redox Biol. 2021, 47, 102150. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Chen, J.; Liu, D.; Zhou, H.; Xiao, W.; Xia, X.; Huang, Z. Cigarette Smoking Is Associated with Human Semen Quality in Synergy with Functional NRF2 Polymorphisms. Biol. Reprod. 2013, 89, 5. [Google Scholar] [CrossRef]
- Masuko, H.; Sakamoto, T.; Kaneko, Y.; Iijima, H.; Naito, T.; Noguchi, E.; Hirota, T.; Tamari, M.; Hizawa, N. An interaction between Nrf2 polymorphisms and smoking status affects annual decline in FEV1: A longitudinal retrospective cohort study. BMC Med. Genet. 2011, 12, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | SNP ID | Study Group | Finding | Reference |
|---|---|---|---|---|
| Nrf2 | rs2364723 | SAPALDIA cohort 2002/2003 (N = 1472) 2010/2011 (N = 1235) | Adverse obesity effects on heart rate variability. | [80] |
| Nrf2 | rs6721961 | Mexican patients: N = 627 diabetic subjects and N = 1020 controls; Chinese patients: N883 T2DM subjects (383 with diabetic nephropathy). | Associated with T2DM development and complications. Higher risk for CAD and severity of coronary atherosclerosis. | [94,95,108] |
| Nrf2 | rs35652124 | Indian subjects N = 400 (100 with diabetic foot ulcer, 150 T2DM patients, and 150 healthy subjects). N = 464 Japanese subjects (285 men and 179 women). N64 healthy African American. N = 184 white individuals | Association with insulin resistance and diabetic complications. Cardiovascular disease-associated mortality. Impairment of vasodilation. | [97,104,122] |
| Nrf2 | rs2364723 | SAPALDIA cohort 2002/2003 (N = 1472) 2010/2011 (N = 1235). N = 1390 subjects from the Vlagtwedde–Vlaardingen cohort. N = 809 Chinese volunteers (214 T2DM patients, 236 without diabetic complications, and 359 healthy individuals). | Impairment of cardiovascular function. Low risk of mortality in cardiovascular diseases. Diabetic complications. Associated with lower triglyceride levels. | [59,80,89] |
| Nrf2 | rs13001694 | N = 1390 subjects from the Vlagtwedde–Vlaardingen cohort. | Associated with low risk of mortality in cardiovascular diseases. | [59] |
| Nrf2 | rs182428269 | N = 400 (150 2DM subjects, 150 healthy subjects, and 100 with diabetic complications). | Risk for T2DM development and diabetic complications | [98] |
| Nrf2 | rs10497511 | N = 809 Chinese volunteers (214 T2DM patients, 236 without complications, and 359 healthy individuals). | Diabetic complications. | [89] |
| Nrf2 | rs1962142 | N = 809 Chinese volunteers (214 T2DM patients, 236 without diabetic complications, and 359 healthy individuals). | Diabetic complications. | [89] |
| Nrf2 | rs6726395 | N = 809 Chinese volunteers (214 T2DM patients, 236 without diabetic complications, and 359 healthy individuals). | Diabetic complications. | [89] |
| KEAP1 | rs11085735 | N = 117 patients with fatal and nonfatal cardiovascular events (42 died). | Predictor of cardiovascular events. | [57] |
| HMOX1 | rs2364723 | N = 809 Chinese volunteers (214 T2DM patients, 236 without diabetic complications, and 359 healthy individuals). | Negative associated with diabetic complications. | [89] |
| GPx | rs1050450 | 416 Mexican women (N = 208 healthy subjects, N = 208 obese subjects). N = 1244 T2DM subjects and N = 730 healthy subjects. N = 773 Caucasian subjects genotyped from the UCL Diabetes and Cardiovascular disease Study and N = 382 Caucasian subjects from the Ealing Diabetes Study. N = 396 T2DM patients and N = 678 control subjects. | Morbid obesity development, particularly in females. Associated with diabetic neuropathy and development of carotid plaques in patients with diabetes. | [90,91,92,93] |
| NQO1 | rs1800566 | N = 2374 Thai subjects (with and without CAD). N = 834 T2DM patients (601 Seoul set and 233 Koyang set). N = 130 patients (67 patients with coronary heart disease and 63 healthy individuals). | Associated with severity of coronary atherosclerosis and CAD in females. Controversial association with cardiovascular risk. Associated with hypertension. | [108,116,117] |
| CAT | rs1049982 | N = 1388 participants > 18 years old (704 women, 300 untreated hypertensive patients). | Risk of hypertension. | [125] |
| TXN | rs2301241 | N = 1388 participants > 18 years old (704 women, 300 untreated hypertensive patients). | Associated with high blood pressure. | [125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zazueta, C.; Jimenez-Uribe, A.P.; Pedraza-Chaverri, J.; Buelna-Chontal, M. Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2. Antioxidants 2022, 11, 507. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11030507
Zazueta C, Jimenez-Uribe AP, Pedraza-Chaverri J, Buelna-Chontal M. Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2. Antioxidants. 2022; 11(3):507. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11030507
Chicago/Turabian StyleZazueta, Cecilia, Alexis Paulina Jimenez-Uribe, José Pedraza-Chaverri, and Mabel Buelna-Chontal. 2022. "Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2" Antioxidants 11, no. 3: 507. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox11030507