Current Concentrations of Zn, Cu, and As in Piggery Wastewater Compromise Nutrient Removals in Microalgae–Bacteria Photobioreactors Due to Altered Microbial Communities
Abstract
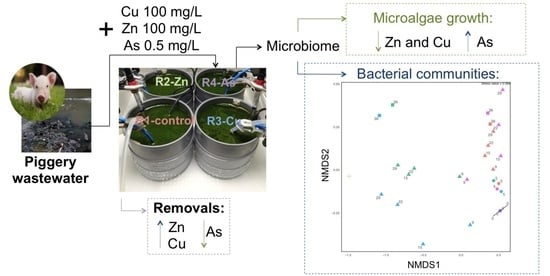
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Piggery Wastewater and Inoculum
2.2. Photobioreactors, Operational Conditions, and Experimental Design
2.3. Genomic DNA Extraction and Amplicon Sequencing
2.4. Statistical Analysis and Diversity
3. Results
3.1. Photobioreactor Performance and Removal of Zn, Cu, and As
3.2. Influence of Zn, Cu, and As on Microalgal Growth
3.3. Bacterial Community Diversity and Structure
3.4. Temporal Changes on the Bacterial Composition of PBRs’ Microbiome
3.5. Effects of the Addition of Metal(loid)s on Bacterial Communities
4. Discussion
4.1. Negative Effect of Zn and Cu on PBRs Performance Associated with Changes in Microbial Communities
4.2. The impact of As on the Removal of Nutrients and the Microbiome of Reactor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PWW | Piggery wastewater |
| HMs | Heavy metals |
| PBRs | Photobioreactors |
| TSS | Total suspended solid |
| TOC | Total organic carbon |
| TN | Total nitrogen |
| NH4+-N | Ammonia nitrogen |
| TP | Total phosphorus |
| HRT | Hydraulic retention time |
| T | Temperature |
| DO | Dissolved oxygen |
| PSII | Photosystem II |
| Fv/Fm | Ratio of variable to maximum fluorescence |
| tbRDA | Transformation-based redundancy analysis |
| NMDS | Non-metric multidimensional scaling |
| SIMPER | Similarity percentages |
References
- Cheng, D.L.; Ngo, H.H.; Guo, W.S.; Chang, S.W.; Nguyen, D.D.; Kumar, S.M. Microalgae Biomass from Swine Wastewater and Its Conversion to Bioenergy. Bioresour. Technol. 2019, 275, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, D.; Kusmayadi, A.; Yen, H.W.; Di Dong, C.; Lee, D.J.; Chang, J.S. Current Advances in Biological Swine Wastewater Treatment Using Microalgae-Based Processes. Bioresour. Technol. 2019, 289, 121718. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zheng, P.; Da, K.; Li, Y.; Li, W.; Dongdong, X.; Chen, W.; Pan, C. The Removal of Copper and Zinc from Swine Wastewater by Anaerobic Biological-Chemical Process: Performance and Mechanism. J. Hazard. Mater. 2021, 401, 123767. [Google Scholar] [CrossRef]
- Zhang, L.; Lee, Y.W.; Jahng, D. Anaerobic Co-Digestion of Food Waste and Piggery Wastewater: Focusing on the Role of Trace Elements. Bioresour. Technol. 2011, 102, 5048–5059. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Forcada, E.; Vega-Alegre, M.; Timón-Sánchez, S. Characterization of Regional Cold-Hydrothermal Inflows Enriched in Arsenic and Associated Trace-Elements in the Southern Part of the Duero Basin (Spain), by Multivariate Statistical Analysis. Sci. Total Environ. 2017, 593–594, 211–226. [Google Scholar] [CrossRef]
- Gao, S.; Hu, C.; Sun, S.; Xu, J.; Zhao, Y.; Zhang, H. Performance of Piggery Wastewater Treatment and Biogas Upgrading by Three Microalgal Cultivation Technologies under Different Initial COD Concentration. Energy 2018, 165, 360–369. [Google Scholar] [CrossRef]
- Nagarajan, D.; Lee, D.J.; Chen, C.Y.; Chang, J.S. Resource Recovery from Wastewaters Using Microalgae-Based Approaches: A Circular Bioeconomy Perspective. Bioresour. Technol. 2020, 302, 122817. [Google Scholar] [CrossRef]
- Fallahi, A.; Rezvani, F.; Asgharnejad, H.; Khorshidi, E.; Hajinajaf, N.; Higgins, B. Interactions of Microalgae-Bacteria Consortia for Nutrient Removal from Wastewater: A Review. Chemosphere 2021, 272, 129878. [Google Scholar] [CrossRef]
- Acién Fernández, F.G.; Gómez-Serrano, C.; Fernández-Sevilla, J.M. Recovery of Nutrients from Wastewaters Using Microalgae. Front. Sustain. Food Syst. 2018, 2, 59. [Google Scholar] [CrossRef] [Green Version]
- García, D.; Posadas, E.; Grajeda, C.; Blanco, S.; Martínez-Páramo, S.; Acién, G.; García-Encina, P.; Bolado, S.; Muñoz, R. Comparative Evaluation of Piggery Wastewater Treatment in Algal-Bacterial Photobioreactors under Indoor and Outdoor Conditions. Bioresour. Technol. 2017, 245, 483–490. [Google Scholar] [CrossRef]
- Cheng, P.; Wang, Y.; Liu, T.; Liu, D. Biofilm Attached Cultivation of Chlorella Pyrenoidosa Is a Developed System for Swine Wastewater Treatment and Lipid Production. Front. Plant Sci. 2017, 8, 1594. [Google Scholar] [CrossRef]
- Suresh Kumar, K.; Dahms, H.U.; Won, E.J.; Lee, J.S.; Shin, K.H. Microalgae—A Promising Tool for Heavy Metal Remediation. Ecotoxicol. Environ. Saf. 2015, 113, 329–352. [Google Scholar] [CrossRef]
- Yang, J.S.; Cao, J.; Xing, G.L.; Yuan, H.L. Lipid Production Combined with Biosorption and Bioaccumulation of Cadmium, Copper, Manganese and Zinc by Oleaginous Microalgae Chlorella Minutissima UTEX2341. Bioresour. Technol. 2015, 175, 537–544. [Google Scholar] [CrossRef]
- Arora, N.; Gulati, K.; Patel, A.; Pruthi, P.A.; Poluri, K.M.; Pruthi, V. A Hybrid Approach Integrating Arsenic Detoxification with Biodiesel Production Using Oleaginous Microalgae. Algal Res. 2017, 24, 29–39. [Google Scholar] [CrossRef]
- Priyadarshanee, M.; Das, S. Biosorption and Removal of Toxic Heavy Metals by Metal Tolerating Bacteria for Bioremediation of Metal Contamination: A Comprehensive Review. J. Environ. Chem. Eng. 2021, 9, 104686. [Google Scholar] [CrossRef]
- Fouilland, E.; Galès, A.; Beaugelin, I.; Lanouguère, E.; Pringault, O.; Leboulanger, C. Influence of Bacteria on the Response of Microalgae to Contaminant Mixtures. Chemosphere 2018, 211, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, C.H.; Lin, M.M.; Ge, Y. A Symbiotic Bacterium Differentially Influences Arsenate Absorption and Transformation in Dunaliella Salina under Different Phosphate Regimes. J. Hazard. Mater. 2016, 318, 443–451. [Google Scholar] [CrossRef]
- De Sousa Oliveira, A.P.; Assemany, P.; Ribeiro Júnior, J.I.; Covell, L.; Nunes-Nesi, A.; Calijuri, M.L. Swine Wastewater Treatment in High Rate Algal Ponds: Effects of Cu and Zn on Nutrient Removal, Productivity and Biomass Composition. J. Environ. Manag. 2021, 299, 113668. [Google Scholar] [CrossRef]
- Li, X.; Yang, C.; Zeng, G.; Wu, S.; Lin, Y.; Zhou, Q.; Lou, W.; Du, C.; Nie, L.; Zhong, Y. Nutrient Removal from Swine Wastewater with Growing Microalgae at Various Zinc Concentrations. Algal Res. 2020, 46, 101804. [Google Scholar] [CrossRef]
- Leong, Y.K.; Chang, J.-S. Bioremediation of Heavy Metals Using Microalgae: Recent Advances and Mechanisms. Bioresour. Technol. 2020, 303, 122886. [Google Scholar] [CrossRef]
- Podder, M.S.; Majumder, C.B. Phycoremediation of Arsenic from Wastewaters by Chlorella Pyrenoidosa. Groundw. Sustain. Dev. 2015, 1, 78–91. [Google Scholar] [CrossRef]
- Zhang, B.; Li, W.; Guo, Y.; Zhang, Z.; Shi, W.; Cui, F.; Lens, P.N.L.; Tay, J.H. Microalgal-Bacterial Consortia: From Interspecies Interactions to Biotechnological Applications. Renew. Sustain. Energy Rev. 2020, 118, 109563. [Google Scholar] [CrossRef]
- Li, H.; Xu, H.; Song, H.L.; Lu, Y.; Yang, X.L. Antibiotic Resistance Genes, Bacterial Communities, and Functions in Constructed Wetland-Microbial Fuel Cells: Responses to the Co-Stresses of Antibiotics and Zinc. Environ. Pollut. 2020, 265, 115084. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Herrera, V.; León, G.; Banihani, Q.; Field, J.A.; Sierra-Alvarez, R. Toxicity of Copper(II) Ions to Microorganisms in Biological Wastewater Treatment Systems. Sci. Total Environ. 2011, 412–413, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Posadas, E.; Alcántara, C.; García-Encina, P.A.; Gouveia, L.; Guieysse, B.; Norvill, Z.; Acién, F.G.; Markou, G.; Congestri, R.; Koreiviene, J.; et al. Microalgae Cultivation in Wastewater. In Microalgae-Based Biofuels and Bioproducts: From Feedstock Cultivation to End-Products; Woodhead Publishing: Thorston, UK, 2017; pp. 67–91. [Google Scholar] [CrossRef]
- Suresh Kumar, K.; Dahms, H.U.; Lee, J.S.; Kim, H.C.; Lee, W.C.; Shin, K.H. Algal Photosynthetic Responses to Toxic Metals and Herbicides Assessed by Chlorophyll a Fluorescence. Ecotoxicol. Environ. Saf. 2014, 104, 51–71. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA). Standards Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association American Water Works Association; Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Saavedra, R.; Muñoz, R.; Taboada, M.E.; Vega, M.; Bolado, S. Comparative Uptake Study of Arsenic, Boron, Copper, Manganese and Zinc from Water by Different Green Microalgae. Bioresour. Technol. 2018, 263, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Sournia, A. Phytoplankton Manual; United Nations Educational, Scientific and Cultural Organization: Paris, France, 1978; Volume 19, ISBN 9231015729. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids. Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.4-6; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. bioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Microbiome Package, version 1.7.21; R/Bioconductor; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Clarke, K.R. Non-Parametric Multivariate Analyses of Changes in Community Structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Collao, J.; del Mar Morales-Amaral, M.; Acién-Fernández, F.G.; Bolado-Rodríguez, S.; Fernandez-Gonzalez, N. Effect of Operational Parameters, Environmental Conditions, and Biotic Interactions on Bacterial Communities Present in Urban Wastewater Treatment Photobioreactors. Chemosphere 2021, 284, 131271. [Google Scholar] [CrossRef]
- Zhou, G.J.; Peng, F.Q.; Zhang, L.J.; Ying, G.G. Biosorption of Zinc and Copper from Aqueous Solutions by Two Freshwater Green Microalgae Chlorella Pyrenoidosa and Scenedesmus Obliquus. Environ. Sci. Pollut. Res. 2012, 19, 2918–2929. [Google Scholar] [CrossRef]
- Liu, H.L.; Chen, B.Y.; Lan, Y.W.; Cheng, Y.C. Biosorption of Zn(II) and Cu(II) by the Indigenous Thiobacillus Thiooxidans. Chem. Eng. J. 2004, 97, 195–201. [Google Scholar] [CrossRef]
- Batool, S.; Hussain, A.; Iqbal, M.A.; Javid, A.; Ali, W.; Bukhari, S.M.; Akmal, M.; Qazi, J.I. Implication of highly metal-resistant microalgal-bacterial co-cultures for the treatment of simulated metal-loaded wastewaters. Int. Microbiol. 2019, 22, 41–48. [Google Scholar] [CrossRef]
- Sekkour, S.; Bekki, A.; Bouchiba, Z.; Vogel, T.M.; Navarro, E. The Diversity of Cultivable Hydrocarbon-Degrading Bacteria Isolated from Crude Oil Contaminated Soil and Sludge from Arzew Refinery in Algeria. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 70–77. [Google Scholar] [CrossRef]
- Zhao, H.P.; Wang, L.; Ren, J.R.; Li, Z.; Li, M.; Gao, H.W. Isolation and Characterization of Phenanthrene-Degrading Strains Sphingomonas sp. ZP1 and Tistrella sp. ZP5. J. Hazard. Mater. 2008, 152, 1293–1300. [Google Scholar] [CrossRef]
- Haberkorn, I.; Walser, J.C.; Helisch, H.; Böcker, L.; Belz, S.; Schuppler, M.; Fasoulas, S.; Mathys, A. Characterization of Chlorella Vulgaris (Trebouxiophyceae) Associated Microbial Communities. J. Phycol. 2020, 56, 1308–1322. [Google Scholar] [CrossRef]
- Ding, P.; Chu, L.; Wang, J. Biological Treatment of Actual Petrochemical Wastewater Using Anaerobic/Anoxic/Oxic Process and the Microbial Diversity Analysis. Appl. Microbiol. Biotechnol. 2016, 100, 10193–10202. [Google Scholar] [CrossRef]
- Ren, T.; Chi, Y.; Wang, Y.; Shi, X.; Jin, X.; Jin, P. Diversified Metabolism Makes Novel Thauera Strain Highly Competitive in Low Carbon Wastewater Treatment. Water Res. 2021, 206, 117742. [Google Scholar] [CrossRef]
- Wang, Q.; He, J. Complete Nitrogen Removal via Simultaneous Nitrification and Denitrification by a Novel Phosphate Accumulating Thauera sp. Strain SND5. Water Res. 2020, 185, 116300. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Huang, H.; Ling, Z.; Yu, Z.; Jiang, Y.; Liu, P.; Li, X. Genome Sequencing Reveals Mechanisms for Heavy Metal Resistance and Polycyclic Aromatic Hydrocarbon Degradation in Delftia Lacustris Strain LZ-C. Ecotoxicology 2016, 25, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tie, B.; Li, Y.; Lei, M.; Wei, X.; Liu, X.; Du, H. Inoculation of Soil with Cadmium-Resistant Bacterium Delftia sp. B9 Reduces Cadmium Accumulation in Rice (Oryza sativa L.) Grains. Ecotoxicol. Environ. Saf. 2018, 163, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Samimi, M.; Shahriari-Moghadam, M. Isolation and Identification of Delftia Lacustris Strain-MS3 as a Novel and Efficient Adsorbent for Lead Biosorption: Kinetics and Thermodynamic Studies, Optimization of Operating Variables. Biochem. Eng. J. 2021, 173, 108091. [Google Scholar] [CrossRef]
- Masoudzadeh, N.; Zakeri, F.; bagheri Lotfabad, T.; Sharafi, H.; Masoomi, F.; Zahiri, H.S.; Ahmadian, G.; Noghabi, K.A. Biosorption of Cadmium by Brevundimonas sp. ZF12 Strain, a Novel Biosorbent Isolated from Hot-Spring Waters in High Background Radiation Areas. J. Hazard. Mater. 2011, 197, 190–198. [Google Scholar] [CrossRef]
- Wu, N.; Xie, S.; Zeng, M.; Xu, X.; Li, Y.; Liu, X.; Wang, X. Impacts of Pile Temperature on Antibiotic Resistance, Metal Resistance and Microbial Community during Swine Manure Composting. Sci. Total Environ. 2020, 744, 140920. [Google Scholar] [CrossRef]
- Zhao, R.; Liu, J.; Feng, J.; Li, X.; Li, B. Microbial Community Composition and Metabolic Functions in Landfill Leachate from Different Landfills of China. Sci. Total Environ. 2021, 767, 144861. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Ciesielski, S.; Korzeniewska, E.; Osińska, A. Environmental Fate of Bacteroidetes, with Particular Emphasis on Bacteroides fragilis Group Bacteria and Their Specific Antibiotic Resistance Genes, in Activated Sludge Wastewater Treatment Plants. J. Hazard. Mater. 2020, 394, 122544. [Google Scholar] [CrossRef]
- Keto, L.; Tsitko, I.; Perttilä, S.; Särkijärvi, S.; Immonen, N.; Kytölä, K.; Alakomi, H.L.; Hyytiäinen-Pabst, T.; Saarela, M.; Rinne, M. Effect of Silage Juice Feeding on Pig Production Performance, Meat Quality and Gut Microbiome. Livest. Sci. 2021, 254, 104728. [Google Scholar] [CrossRef]
- Jia, L.; Jiang, B.; Huang, F.; Hu, X. Nitrogen Removal Mechanism and Microbial Community Changes of Bioaugmentation Subsurface Wastewater Infiltration System. Bioresour. Technol. 2019, 294, 122140. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, P.; He, Y.; Zeng, F.; Xu, J.; He, L. Enantioselective Effects of Cyflumetofen on Microbial Community and Related Nitrogen Cycle Gene Function in Acid-Soil. Sci. Total Environ. 2021, 771, 144831. [Google Scholar] [CrossRef]
- Frigon, D.; Wells, G. Microbial Immigration in Wastewater Treatment Systems: Analytical Considerations and Process Implications. Curr. Opin. Biotechnol. 2019, 57, 151–159. [Google Scholar] [CrossRef]
- Araújo, E.; Strawn, D.G.; Morra, M.; Moore, A.; Ferracciú Alleoni, L.R. Association between Extracted Copper and Dissolved Organic Matter in Dairy-Manure Amended Soils. Environ. Pollut. 2019, 246, 1020–1026. [Google Scholar] [CrossRef]
- Torres-Franco, A.F.; Figueredo, C.C.; Barros, L.; Gücker, B.; Boëchat, I.G.; Muñoz, R.; Mota, C.R. Assessment of a Deep, LED-Enhanced High-Rate Algal Pond for the Treatment of Digestate. Algal Res. 2021, 59, 102444. [Google Scholar] [CrossRef]
- Lu, H.; Chandran, K.; Stensel, D. Microbial Ecology of Denitrification in Biological Wastewater Treatment. Water Res. 2014, 64, 237–254. [Google Scholar] [CrossRef]
- Kim, S.J.; Ahn, J.H.; Lee, T.H.; Weon, H.Y.; Hong, S.B.; Seok, S.J.; Whang, K.S.; Kwon, S.W. Reyranella soli sp. nov., Isolated from Forest Soil, and Emended Description of the Genus Reyranella Pagnier et al. 2011. Int. J. Syst. Evol. Microbiol. 2013, 63, 3164–3167. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xu, W.; Yu, J.; Zhang, A.; Peng, F.; Lian, J.; Huang, J.; Pang, Q.; Zhu, X.; Gao, P. Response of Denitrobacteria Involved in Nitrogen Removal for Treatment of Simulated Livestock Wastewater Using a Novel Bioreactor. Ecol. Eng. 2020, 147, 105762. [Google Scholar] [CrossRef]
- He, T.; Xie, D.; Ni, J.; Cai, X.; Li, Z. Investigating the Effect of Copper and Magnesium Ions on Nitrogen Removal Capacity of Pure Cultures by Modified Non-Competitive Inhibition Model. Ecotoxicol. Environ. Saf. 2019, 170, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.R.; Wang, Y.; Chen, H.; Lyu, Y.K. Ammonium Removal Characteristics of an Acid-Resistant Bacterium Acinetobacter sp. JR1 from Pharmaceutical Wastewater Capable of Heterotrophic Nitrification-Aerobic Denitrification. Bioresour. Technol. 2019, 274, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, F.; Liu, Y.; Huang, F.; Zhang, C. Effect of Extracellular Polymeric Substances on Arsenic Accumulation in Chlorella Pyrenoidosa. Sci. Total Environ. 2020, 704, 135368. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, S.; Xu, P.; Liu, C.; Liu, M.; Wang, Y.; Wang, C.; Zhang, C.; Ge, Y. Review of Arsenic Speciation, Toxicity and Metabolism in Microalgae. Rev. Environ. Sci. Biotechnol. 2015, 14, 427–451. [Google Scholar] [CrossRef]
- Bahar, M.M.; Megharaj, M.; Naidu, R. Influence of Phosphate on Toxicity and Bioaccumulation of Arsenic in a Soil Isolate of Microalga Chlorella sp. Environ. Sci. Pollut. Res. 2016, 23, 2663–2668. [Google Scholar] [CrossRef]
- Saavedra, R.; Muñoz, R.; Taboada, M.E.; Bolado, S. Influence of Organic Matter and CO2 Supply on Bioremediation of Heavy Metals by Chlorella Vulgaris and Scenedesmus Almeriensis in a Multimetallic Matrix. Ecotoxicol. Environ. Saf. 2019, 182, 109393. [Google Scholar] [CrossRef] [Green Version]
- Nazina, T.; Babich, T.; Kostryukova, N.; Sokolova, D.; Abdullin, R.; Tourova, T.; Kadnikov, V.; Mardanov, A.; Ravin, N.; Grouzdev, D.; et al. Ultramicrobacteria from Nitrate-and Radionuclide-Contaminated Groundwater. Sustainability 2020, 12, 1239. [Google Scholar] [CrossRef] [Green Version]
- Paddock, M.B.; Fernández-Bayo, J.D.; VanderGheynst, J.S. The Effect of the Microalgae-Bacteria Microbiome on Wastewater Treatment and Biomass Production. Appl. Microbiol. Biotechnol. 2020, 104, 893–905. [Google Scholar] [CrossRef]
- Durante-Rodríguez, G.; Fernández-Llamosas, H.; Alonso-Fernandes, E.; Fernández-Muñiz, M.N.; Muñoz-Olivas, R.; Díaz, E.; Carmona, M. ArxA from Azoarcus sp. CIB, an Anaerobic Arsenite Oxidase from an Obligate Heterotrophic and Mesophilic Bacterium. Front. Microbiol. 2019, 10, 1699. [Google Scholar] [CrossRef] [Green Version]
- Papirio, S.; Zou, G.; Ylinen, A.; Di Capua, F.; Pirozzi, F.; Puhakka, J.A. Effect of Arsenic on Nitrification of Simulated Mining Water. Bioresour. Technol. 2014, 164, 149–154. [Google Scholar] [CrossRef]
- Deng, F.; Zhang, D.; Yang, L.; Li, L.; Lu, Y.; Wang, J.; Fan, Y.; Zhu, Y.; Li, X.; Zhang, Y. Effects of Antibiotics and Heavy Metals on Denitrification in Shallow Eutrophic Lakes. Chemosphere 2022, 291, 132948. [Google Scholar] [CrossRef]
- Ma, J.J.; Jiang, C.L.; Tao, X.H.; Sheng, J.L.; Sun, X.Z.; Zhang, T.Z.; Zhang, Z.J. Insights on Dissolved Organic Matter and Bacterial Community Succession during Secondary Composting in Residue after Black Soldier Fly Larvae (Hermetia illucens L.) Bioconversion for Food Waste Treatment. Waste Manag. 2022, 142, 55–64. [Google Scholar] [CrossRef]
- Aguilar, N.C.; Faria, M.C.S.; Pedron, T.; Batista, B.L.; Mesquita, J.P.; Bomfeti, C.A.; Rodrigues, J.L. Isolation and Characterization of Bacteria from a Brazilian Gold Mining Area with a Capacity of Arsenic Bioaccumulation. Chemosphere 2020, 240, 124871. [Google Scholar] [CrossRef]
- Krustok, I.; Truu, J.; Odlare, M.; Truu, M.; Ligi, T.; Tiirik, K.; Nehrenheim, E. Effect of Lake Water on Algal Biomass and Microbial Community Structure in Municipal Wastewater-Based Lab-Scale Photobioreactors. Appl. Microbiol. Biotechnol. 2015, 99, 6537–6549. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Q.; Ren, H.; Min, M.; Chen, P.; Ruan, R. Isolation of a Bacterial Strain, Acinetobacter sp. from Centrate Wastewater and Study of Its Cooperation with Algae in Nutrients Removal. Bioresour. Technol. 2017, 235, 59–69. [Google Scholar] [CrossRef]





| Parameters (Unit) | Experiment A | Experiment B |
|---|---|---|
| Total suspended solid (TSS in g/L) | 0.42 ± 0.1 | 0.3 ± 0.02 |
| Total organic carbon (TOC in mg/L) | 713 ± 26 | 652 ± 27 |
| Total nitrogen (TN in mg/L) | 256 ± 11 | 247 ± 11 |
| Ammonia nitrogen (NH4+-N in mg/L) | 141 ± 6 | 164 ± 10 |
| Total phosphorus (TP in mg/L) | 3.8 ± 0.21 | 3.6 ± 0.17 |
| Zinc (Zn in mg/L) | 0.98 ± 0.63 | 0.78 ± 0.23 |
| Copper (Cu in mg/L) | 0.28 ± 0.15 | 0.25 ± 0.05 |
| Arsenic (As in μg/L) | 2.04 ± 0.19 | 1.94 ± 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collao, J.; García-Encina, P.A.; Blanco, S.; Bolado-Rodríguez, S.; Fernandez-Gonzalez, N. Current Concentrations of Zn, Cu, and As in Piggery Wastewater Compromise Nutrient Removals in Microalgae–Bacteria Photobioreactors Due to Altered Microbial Communities. Biology 2022, 11, 1176. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11081176
Collao J, García-Encina PA, Blanco S, Bolado-Rodríguez S, Fernandez-Gonzalez N. Current Concentrations of Zn, Cu, and As in Piggery Wastewater Compromise Nutrient Removals in Microalgae–Bacteria Photobioreactors Due to Altered Microbial Communities. Biology. 2022; 11(8):1176. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11081176
Chicago/Turabian StyleCollao, Javiera, Pedro Antonio García-Encina, Saúl Blanco, Silvia Bolado-Rodríguez, and Nuria Fernandez-Gonzalez. 2022. "Current Concentrations of Zn, Cu, and As in Piggery Wastewater Compromise Nutrient Removals in Microalgae–Bacteria Photobioreactors Due to Altered Microbial Communities" Biology 11, no. 8: 1176. https://0-doi-org.brum.beds.ac.uk/10.3390/biology11081176