Caffeic Acid Esters Are Effective Bactericidal Compounds Against Paenibacillus larvae by Altering Intracellular Oxidant and Antioxidant Levels
Abstract
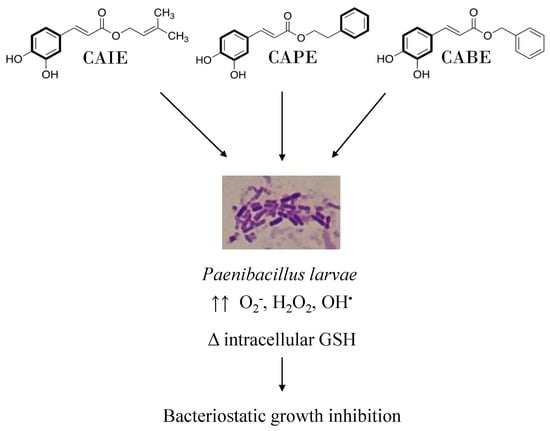
:1. Introduction
2. Materials and Methods
2.1. General Materials for Synthetic Procedures
2.2. Preparation of Caffeic Ester Derivatives
2.2.1. Caffeic Acid Cinnamyl Ester Diacetate
2.2.2. Caffeic Acid Benzyl Ester Diacetate
2.2.3. Caffeic Acid Phenethyl Ester Diacetate
2.2.4. Caffeic Acid Isopropenyl Ester Diacetate
2.2.5. Caffeic Acid Cinnamyl Ester (CACE)
2.2.6. Caffeic Acid Benzyl Ester
2.2.7. Caffeic Acid Phenethyl Ester
2.2.8. Caffeic Acid Isopropenyl Ester
2.3. Bacterial Strains and Growth Conditions
2.4. Determination of Minimal Inhibitory Concentration and Minimal Bactericidal Concentration
2.5. Flow Cytometry Analysis
2.6. Quantification of Intracellular Reactive Oxygen Species and Glutathione Levels
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bruckner, S.; Steinhauer, N.; Rennich, K.; Aurell, D.; Caron, D.; Ellis, J.; Marie Fauvel, A.; Kulhanek, K.; Nelson, K.; Rangel, J.; et al. Honey Bee Colony Losses 2017–2018: Preliminary Results. Available online: https://beeinformed.org/results/honey-bee-colony-losses-2017–2018-preliminary-results/ (accessed on 15 February 2019).
- Goulson, D.; Nicholls, E.; Botias, C.; Ratheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, N.; Kulhanek, K.; Antunez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.; van Engelsdorp, D. Drivers of colony losses. Curr. Opin. Insect Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Nazzi, F.; Brown, S.; Annoscia, D.; Del Piccolo, F.; Varricchio, P. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathogen. 2012, 8, e1002735. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E. American foulbrood in honeybees and its caustitive agent, Paenibacillus larvae. J. Invert. Pathol. 2010, 103, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Djukic, M.; Brzuszkiewicz, E.; Funfhaus, A.; Voss, J.; Gollnow, K.; Poppinga, L.; Liesegang, H.; Garcia-Gonzales, E.; Genersch, E.; Daniel, R. How to kill the honey bee larva: Genomic potential and virulence mechanisms of Paenibacillus larvae. PLoS ONE 2014, 3, e90914. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.; Brodsgaard, C. American foulbrood: A review of its biology, diagnosis and control. Bee World 1999, 80, 5–23. [Google Scholar] [CrossRef]
- Ansari, M.; Al-Ghamdi, A.; Nuru, A.; Ahmed, A.; Ayaad, T.; Khan, K.; Al-Waili, N. Diagnosis and molecular detection of Paenibacilus larvae, the causative agent of American foulbrood in honeybees in Saudi Arabia. Int. J. Trop. Insect. Sci. 2017, 37, 137–148. [Google Scholar] [CrossRef]
- Cornman, R.; Lopez, D.; Evans, J. Transcriptional response of honey bee larvae infected with the bacterial pathogen Paenibacillus larvae. PLoS ONE 2013, 8, e65424. [Google Scholar] [CrossRef]
- Powell, J.; Martinson, V.; Urban-Mead, K.; Moran, N. Routes of acquisition of the gut microbiota of the honey bee Apis melifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- De Graaf, D.; Alippa, A.; Brown, M.; Evans, J.; Feldlaufer, M. Diagnosis of American foulbrood in honey bees: A synthesis and proposed analytical protocols. Lett. Appl. Microbiol. 2006, 43, 583–590. [Google Scholar] [CrossRef]
- Kochansky, J.; Knox, D.; Feldlaufer, M.; Pettis, J. Screening alternative antibiotics against oxytetracycline susceptible and resistant Paenibacillus larvae. Apidologie 2001, 32, 215–222. [Google Scholar] [CrossRef]
- Mutinelli, F. European legislation governing the authorization of veterinary medicinal products with particular reference to the use of drugs for the control of honey bee diseases. Apiacta 2003, 38, 156–168. [Google Scholar] [CrossRef]
- Sanford, M. Antibiotics and the Beekeeper–The FDA’s Veterinarian Feed Directive (VFD). American Bee Journal. Available online: https://americanbeejournal.com/antibiotics-and-the-beekeeper, (accessed on 15 February 2019).
- Simone-Finstrom, M.; Spivak, M. Increased resin collection after parasite challenge: A case of self-medication in honey bees? PLoS ONE 2012, 7, e34061. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M. Social immunity and the superorganism: Behavioral defenses protecting honey bee colonies from pathogens and parasites. Bee World 2017, 94, 21–29. [Google Scholar] [CrossRef]
- Evans, J.; Spivak, M. Socialized medicine: Individual and communal disease barriers in honey bees. J. Invert. Path. 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Parasites in Social Insects; Princeton University Press: Princeton, NJ, USA, 1998. [Google Scholar]
- Simone, M.; Evans, J.; Spivak, M. Resin collection and social immunity in honey bees. Evolution 2009, 63, 2016–3022. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M.; Spivak, M. Propolis and bee health: The natural history and significance of resin use by honey bees. Apidologie 2010, 41, 295–311. [Google Scholar] [CrossRef]
- Visscher, P. Adaptations of honey bees (Apis millifera) to problems of nest hygiene. Sociobiology 1980, 5, 249–260. [Google Scholar]
- Lagenheim, J. Plant. Resins: Chemistry, Evolution, Ecology, and Ethanobotany; Timber Press: Portland, OR, USA, 2003. [Google Scholar]
- Burdock, G. Review of the biological properties and toxicity of bee propolis (propolis). Food Chem. Toxicol. 1998, 36, 347–363. [Google Scholar] [CrossRef]
- Macucci, M. Propolis: Chemical composition, biological properties and therapeutic activity. Apidologie 1995, 26, 83–99. [Google Scholar] [CrossRef]
- Alonso-Salces, R.; Cugnata, N.; Guaspari, E.; Pellegrini, M.; Aubone, I.; Antunez, K.; Fuselli, S. Natural strategies for the control of Paenibacillus larvae, the causative agent of American foulbrood in honey bees: A review. Apidologie 2017, 48, 387–400. [Google Scholar] [CrossRef]
- Wilson, M.; Brinkman, D.; Spivak, M.; Gardner, G.; Cohen, J. Regional variation in composition and antimicrobial activity of US propolis against Paenibacillus larvae and Ascophaera apis. J. Invert. Path. 2015, 124, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Bastos, E.; Simone, M.; Macedo Jorge, D.; Egea Soares, A.; Spivak, M. In vitro study of the antimicrobial activity of Brazilian propolis against Paenibacillus larvae. J. Invert. Path. 2008, 97, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Siedel, V.; Peyfoon, E.; Watson, D.; Fernley, J. Comparative study of the antibacterial activity of propolis from different geographical and climatic zones. Phytotherapy Res. 2008, 22, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Sforcin, J.; Bankova, V. Propolis: Is there a potential for the development of new drugs? J. Ethanopharm. 2011, 133, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Woisky, R.; Salatino, A. Analysis of propolis: Some parameters and procedures for chemical quality control. J. Apic. Res. 1998, 37, 99–105. [Google Scholar] [CrossRef]
- Guzman, J. Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 2014, 19, 19292–19349. [Google Scholar] [CrossRef]
- Meyuhas, S.; Assali, M.; Huleihil, M. Antimicrobial activities of caffeic acid phenethyl ester. J. Molec. Biochem. 2015, 4, 21–31. [Google Scholar]
- Bilikova, K.; Papova, M.; Trusheva, B.; Bankova, V. New anti-Paenibacillus larvae substances purified from propolis. Apidologie 2013, 44, 278–285. [Google Scholar] [CrossRef]
- Alippi, A. American Foulbrood of Honey Bees (Infection of Honey Bees with Paenibacillus larvae). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organisation for Animal Health: Paris, France, 2016; pp. 1–17. [Google Scholar]
- Malmberg, C. Evaluation of flow Cytometry as Replacement for Plating in in vitro Measurements of Competitive Growth under Antibiotic Stress; Department of Infectious Diseases. Uppsala University: Uppsala, Sweden, 2010. [Google Scholar]
- Van Acker, H.; Gielis, M.; Acke, F.; Cools, P.; Cos, T. The role of reactive oxygen species in antibioitic-induced cell death in Burkholderia cepacia complex bacteria. PLoS ONE 2016, 11, e0159837. [Google Scholar] [CrossRef]
- LeBlanc, L.; Pare, A.; Jean-Francois, J.; Hebert, M.; Surette, M.; Touaibia, M. Synthesis and antiradical/antioxidant activities of caffeic acid phenethyl ester and its related propionic, acetic and benzoic acid analogues. Molecules 2012, 17, 14637–14650. [Google Scholar] [CrossRef] [PubMed]
- Bankova, V.; Trusheva, B.; Popova, M. Caffeic Acid Phenethyl Ester (CAPE)–NATURAL Sources, Analytical Procedures and Synthetic Approaches; Comptes rendus de l’Académie bulgare des sciences; Sciences mathématiques et naturelles: Sofia, Bulgaria, 2018; Volume 71, pp. 1157–1169. [Google Scholar]
- Jayaprakasam, B.; Vanisree, M.; Zhang, Y.; Dewitt, D.; Nair, M. Impact of alkyl esters of caffeic and ferulic acids on tumor cell proliferation, cyclooxygenase enzyme, and lipid peroxidation. J. Agric. Food Chem. 2006, 54, 5375–5381. [Google Scholar] [CrossRef]
- Schweigert, N.; Zehnder, A.; Eggen, R. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 2001, 3, 81–91. [Google Scholar] [CrossRef]
- Schweigert, N.; Acero, J.; von Gunten, U.; Canonica, S.; Zehnder, A.; Eggen, R. DNA degradation by the mixture of copper and catechol is caused by DNA-copper oxo complexes, probably DNA-Cu (I) OOH. Environ. Mol. Mutagen. 2000, 36, 5–12. [Google Scholar] [CrossRef]
- Zhang, P.; Tang, Y.; Li, N.; Zhu, Y.; Duan, J. Bioactivity and chemical synthesis of caffeic acid phenethyl ester and its derivatives. Molecules 2014, 19, 16458–16476. [Google Scholar] [CrossRef]
- Banskota, A.; Tezuka, Y.; Kadota, S. Recent progress in pharmacological research of propolis. Phytother. Res. 2001, 15, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Cueva, C.; Moreno-Arribas, M.; Martin-Alvarez, P.; Bills, G.; Vicente, M.; Basilio, A.; Rivas, C.; Requena, T.; Rodriguez, J.; Bartolome, B. Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res. Microbiol. 2010, 161, 372–382. [Google Scholar] [CrossRef]
- Flesar, J.; Havlik, J.; Kloucek, P.; Rada, V.; Titera, D.; Bednarm, M.; Stropnicky Mand Kokoska, L. In vitro growth-inhibitory effect of plant-derived extracts and compounds against Paenibacillus larvae and their acute oral toxicity to adult honey bees. Vet. Microbiol. 2010, 145, 129–133. [Google Scholar] [CrossRef]
- Merkl, R.; Hradkova, L.; Filip, V.; Smidrkal, J. Antimicrobial and antioxidant properties of phenolic acids alkyl esters. Czech J. Food Sci. 2010, 28, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Chiao, C.; Carothers, A.; Grunberger, D.; Solomon, G.; Preston, G.; Barrett, J. Apoptosis and altered redox state induced by caffeic acid phenethyl ester (CAPE) in transformed rat fibroblast cells. Cancer Res. 1995, 55, 3576–3583. [Google Scholar]
- Blake, D.J.; Bolin, C.M.; Cox, D.P.; Cardozo-Palaez, F.; Pfau, J.C. Internalization of Libby amphibole asbestos and induction of oxidative stress in murine macrophages. Toxicol. Sci. 2007, 99, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J. Diagnosing oxidative stress in bacteria: Not as easy as you might think. Curr. Opin. Microbiol. 2015, 24, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Meng, D.; Wang, Z.; Guo, H.; Wang, Y. Oxidative stress response in two representative bacteria exposed to atrazine. FEMS Microbiol. Lett. 2012, 334, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar] [PubMed]




| Compound Name | Compound Abbreviation | Molecular Weight | MIC (µg/mL) | MBC (µg/mL) |
|---|---|---|---|---|
| Caffeic Acid | CA | 180.16 | >500 | ND |
| Ferulic Acid | FA | 194.19 | >500 | ND |
| Caffeic acid cinnamyl ester | CACE | 296.32 | >500 | ND |
| Caffeic acid isopropenyl ester | CAIE | 248.28 | 125 | 125 |
| Caffeic acid phenethyl ester | CAPE | 284.31 | 125 | 125 |
| Caffeic acid benzyl ester | CABE | 270.28 | 125 | 125 |
| Combination of 3 compounds | CAIE, CAPE, CABE | 31.25 | 31.25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collins, W.; Lowen, N.; Blake, D.J. Caffeic Acid Esters Are Effective Bactericidal Compounds Against Paenibacillus larvae by Altering Intracellular Oxidant and Antioxidant Levels. Biomolecules 2019, 9, 312. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080312
Collins W, Lowen N, Blake DJ. Caffeic Acid Esters Are Effective Bactericidal Compounds Against Paenibacillus larvae by Altering Intracellular Oxidant and Antioxidant Levels. Biomolecules. 2019; 9(8):312. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080312
Chicago/Turabian StyleCollins, William, Noah Lowen, and David J. Blake. 2019. "Caffeic Acid Esters Are Effective Bactericidal Compounds Against Paenibacillus larvae by Altering Intracellular Oxidant and Antioxidant Levels" Biomolecules 9, no. 8: 312. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080312